1H1Y
 
 | |
1GY0
 
 | |
1H6J
 
 | |
1GRH
 
 | | INHIBITION OF HUMAN GLUTATHIONE REDUCTASE BY THE NITROSOUREA DRUGS 1,3-BIS(2-CHLOROETHYL)-1-NITROSOUREA AND 1-(2-CHLOROETHYL)-3-(2-HYDROXYETHYL)-1-NITROSOUREA | | Descriptor: | ETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, ... | | Authors: | Karplus, P.A, Schulz, G.E. | | Deposit date: | 1992-12-15 | | Release date: | 1994-01-31 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of human glutathione reductase by the nitrosourea drugs 1,3-bis(2-chloroethyl)-1-nitrosourea and 1-(2-chloroethyl)-3-(2-hydroxyethyl)-1-nitrosourea. A crystallographic analysis.
Eur.J.Biochem., 171, 1988
|
|
1H3G
 
 | | Cyclomaltodextrinase from Flavobacterium sp. No. 92: from DNA sequence to protein structure | | Descriptor: | CALCIUM ION, Cyclomaltodextrinase | | Authors: | Fritzsche, H.B, Schwede, T, Jelakovic, S, Schulz, G.E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-08-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Covalent and Three-Dimensional Structure of the Cyclodextrinase from Flavobacterium Sp. No. 92.
Eur.J.Biochem., 270, 2003
|
|
1GXZ
 
 | |
1H6S
 
 | |
1H1Z
 
 | |
2P6P
 
 | |
2V03
 
 | | High resolution structure and catalysis of an O-acetylserine sulfhydrylase | | Descriptor: | CITRIC ACID, Cysteine synthase B, GLYCEROL, ... | | Authors: | Zocher, G, Wiesand, U, Schulz, G.E. | | Deposit date: | 2007-05-08 | | Release date: | 2007-10-09 | | Last modified: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High resolution structure and catalysis of O-acetylserine sulfhydrylase isozyme B from Escherichia coli.
FEBS J., 274, 2007
|
|
2VKD
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF LETHAL TOXIN FROM CLOSTRIDIUM SORDELLII IN COMPLEX WITH UDP-GLC AND MANGANESE ION | | Descriptor: | CYTOTOXIN L, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ziegler, M.O.P, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Conformational Changes and Reaction of Clostridial Glycosylating Toxins.
J.Mol.Biol., 377, 2008
|
|
2VK9
 
 | |
2VKH
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF LETHAL TOXIN FROM CLOSTRIDIUM SORDELLII IN COMPLEX WITH UDP-GLC AND CALCIUM ION | | Descriptor: | CALCIUM ION, CYTOTOXIN L, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ziegler, M.O.P, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2007-12-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Changes and Reaction of Clostridial Glycosylating Toxins.
J.Mol.Biol., 377, 2008
|
|
2VL8
 
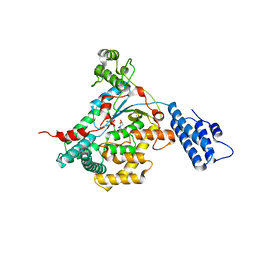 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF LETHAL TOXIN FROM CLOSTRIDIUM SORDELLII IN COMPLEX WITH UDP, CASTANOSPERMINE AND CALCIUM ION | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, CYTOTOXIN L, ... | | Authors: | Jank, T, Ziegler, M.O.P, Schulz, G.E, Aktories, K. | | Deposit date: | 2008-01-09 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Inhibition of the Glucosyltransferase Activity of Clostridial Rho/Ras-Glucosylating Toxins by Castanospermine.
FEBS Lett., 582, 2008
|
|
2VQH
 
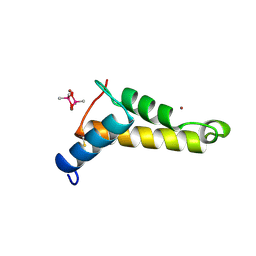 | |
2VQK
 
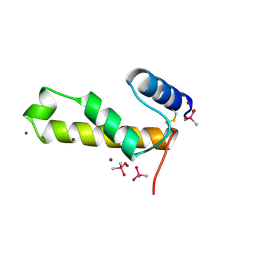 | |
2VQL
 
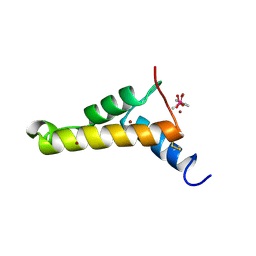 | |
2VSA
 
 | | Structure and mode of action of a mosquitocidal holotoxin | | Descriptor: | MOSQUITOCIDAL TOXIN | | Authors: | Treiber, N, Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure and Mode of Action of a Mosquitocidal Holotoxin
J.Mol.Biol., 381, 2008
|
|
2VQG
 
 | | Crystal structure of PorB from Corynebacterium glutamicum (crystal form I) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 3,6,9,12,15,18,21,24,27-nonaoxaheptatriacontan-1-ol, ACETATE ION, ... | | Authors: | Ziegler, K, Benz, R, Schulz, G.E. | | Deposit date: | 2008-03-15 | | Release date: | 2008-05-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A Putative Alpha-Helical Porin from Corynebacterium Glutamicum.
J.Mol.Biol., 379, 2008
|
|
2VOU
 
 | | Structure of 2,6-dihydroxypyridine-3-hydroxylase from Arthrobacter nicotinovorans | | Descriptor: | 2,6-DIHYDROXYPYRIDINE HYDROXYLASE, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Treiber, N, Schulz, G.E. | | Deposit date: | 2008-02-21 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of 2,6-Dihydroxypyridine 3-Hydroxylase from a Nicotine-Degrading Pathway.
J.Mol.Biol., 379, 2008
|
|
2VSE
 
 | | Structure and mode of action of a mosquitocidal holotoxin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, MOSQUITOCIDAL TOXIN | | Authors: | Treiber, N, Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mode of Action of a Mosquitocidal Holotoxin.
J.Mol.Biol., 381, 2008
|
|
4GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, MIXED DISULFIDE BETWEEN TRYPANOTHIONE AND THE ENZYME | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
5GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, GLUTATHIONYLSPERMIDINE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, GLUTATHIONYLSPERMIDINE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
3GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED TRYPANOTHIONE COMPLEX | | Descriptor: | 2-AMINO-4-[4-(4-AMINO-4-CARBOXY-BUTYRYLAMINO)-5,8,19,22-TETRAOXO-1,2-DITHIA-6,9,13,18,21-PENTAAZA-CYCLOTETRACOS-23-YLCARBAMOYL]-BUTYRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
2GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED GLUTATHIONE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
