3UP6
 
 | |
1GIB
 
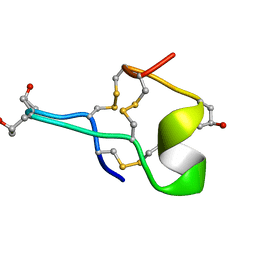 | | MU-CONOTOXIN GIIIB, NMR | | Descriptor: | MU-CONOTOXIN GIIIB | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels.
Biochemistry, 35, 1996
|
|
2ZK9
 
 | | Crystal Structure of Protein-glutaminase | | Descriptor: | GLYCEROL, Protein-glutaminase, SODIUM ION | | Authors: | Hashizume, R. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-17 | | Last modified: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of protein glutaminase and its pro forms converted into enzyme-substrate complex
J.Biol.Chem., 286, 2011
|
|
3A56
 
 | |
3A55
 
 | |
3A54
 
 | |
3WV2
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydroquinazoline-2-carboxamide | | Descriptor: | CALCIUM ION, Collagenase 3, N-(3-methoxybenzyl)-4-oxo-3,4-dihydroquinazoline-2-carboxamide, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
3WV3
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
3WV1
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with 4-(2-((6-fluoro-2-((3-methoxybenzyl)carbamoyl)-4-oxo-3,4-dihydroquinazolin-5-yl)oxy)ethyl)benzoic acid | | Descriptor: | 4-[2-({6-fluoro-2-[(3-methoxybenzyl)carbamoyl]-4-oxo-3,4-dihydroquinazolin-5-yl}oxy)ethyl]benzoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of novel, highly potent, and selective quinazoline-2-carboxamide-based matrix metalloproteinase (MMP)-13 inhibitors without a zinc binding group using a structure-based design approach
J.Med.Chem., 57, 2014
|
|
2EC7
 
 | | Solution Structure of Human Immunodificiency Virus Type-2 Nucleocapsid Protein | | Descriptor: | Gag polyprotein (Pr55Gag), ZINC ION | | Authors: | Matsui, T, Kodera, Y, Tanaka, T, Endoh, H, Tanaka, H, Miyauchi, E, Komatsu, H, Kohno, T, Maeda, T. | | Deposit date: | 2007-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RNA recognition mechanism of human immunodeficiency virus (HIV) type 2 NCp8 is different from that of HIV-1 NCp7
Biochemistry, 48, 2009
|
|
3VW3
 
 | | Antibody 64M-5 Fab in complex with a double-stranded DNA (6-4) photoproduct | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), COBALT HEXAMMINE(III), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y. | | Deposit date: | 2012-07-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a double-stranded DNA (6-4) photoproduct in complex with the 64M-5 antibody Fab
Acta Crystallogr.,Sect.D, 69, 2013
|
|
