3B21
 
 | |
4XZX
 
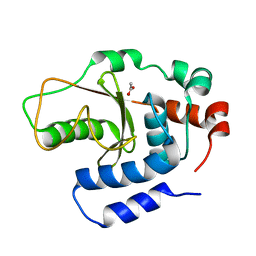 | | Shigella flexneri effector OspI C62S mutant | | Descriptor: | ACETATE ION, ORF169b | | Authors: | Nishide, A, Takagi, K, Minsoo, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New insights into the active site structure of Shigella effecter OspI
To Be Published
|
|
3L3P
 
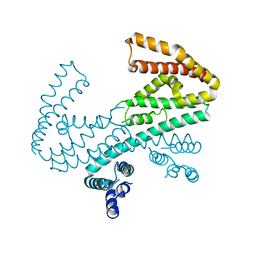 | | Crystal structure of the C-terminal domain of Shigella type III effector IpaH9.8, with a novel domain swap | | Descriptor: | IpaH9.8 | | Authors: | Seyedarabi, A, Sullivan, J.A, Sasakawa, C, Pickersgill, R.W. | | Deposit date: | 2009-12-17 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A disulfide driven domain swap switches off the activity of Shigella IpaH9.8 E3 ligase
Febs Lett., 584, 2010
|
|
5B0N
 
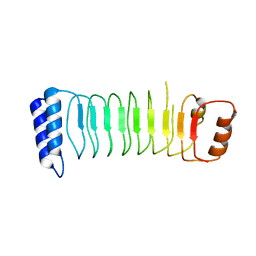 | | Structure of Shigella effector LRR domain | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Takagi, K, Sasakawa, C, Kim, M, Mizushima, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the substrate-recognition domain of the Shigella E3 ligase IpaH9.8
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B0T
 
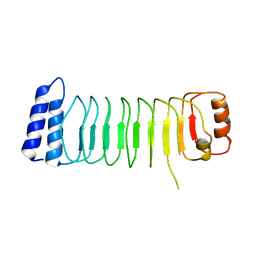 | | Structure of Shigella effector LRR domain | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Takagi, K, Sasakawa, C, Kim, M, Mizushima, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the substrate-recognition domain of the Shigella E3 ligase IpaH9.8
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3W31
 
 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | IODIDE ION, ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
J.Mol.Biol., 425, 2013
|
|
3W30
 
 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | ORF169b | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Basis for the Recognition of Ubc13 by the Shigella flexneri Effector OspI.
J.Mol.Biol., 425, 2013
|
|
8P5P
 
 | | Structure of TECPR1 N-terminal DysF domain | | Descriptor: | GLYCEROL, Tectonin beta-propeller repeat-containing protein 1 | | Authors: | Boyle, K.B, Elliott, P.R, Randow, F. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TECPR1 conjugates LC3 to damaged endomembranes upon detection of sphingomyelin exposure.
Embo J., 42, 2023
|
|
