7DJQ
 
 | |
7YOK
 
 | | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A | | Descriptor: | Cysteine synthase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A
To Be Published
|
|
7YOM
 
 | | Crystal structure of tetra mutant (D67E,A68P,L98I,A301S) of O-acetylserine sulfhydrylase from Salmonella typhimurium in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.8 A | | Descriptor: | Cysteine synthase, peptide from serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of I88L single mutant of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.14 A
To Be Published
|
|
7YOL
 
 | | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide of serine acetyltransferase from Haemophilus influenzae at 2.4 A | | Descriptor: | Cysteine synthase, Peptide from Serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Haemophilus influenzae at 2.4 A
To Be Published
|
|
5XOQ
 
 | |
7EWO
 
 | | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase | | Authors: | Rahisuddin, R, Ekka, M.K, Singh, A.K, Saini, N, Patel, M, Kumar, N, Kumaran, S. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae
To Be Published
|
|
7CM8
 
 | | High resolution crystal structure of M92A mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase, GLYCEROL, SODIUM ION | | Authors: | Kaushik, A, Rahisuddin, R, Saini, N, Kumaran, S. | | Deposit date: | 2020-07-25 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of selective substrate engagement and inhibitor disengagement of cysteine synthase.
J.Biol.Chem., 296, 2020
|
|
7YOE
 
 | |
7YOF
 
 | |
7YOG
 
 | |
7YOD
 
 | |
7YOH
 
 | |
7YOI
 
 | |
7C35
 
 | |
5XCW
 
 | |
5XCP
 
 | |
5XCN
 
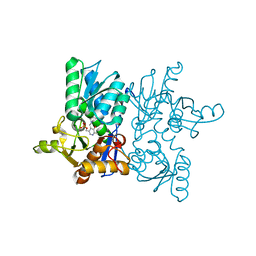 | |
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6TNU
 
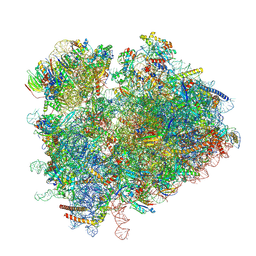 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
