6PL6
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, ... | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6PL5
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
8T9N
 
 | | Bacillus subtilis RsgI GGG mutant | | Descriptor: | Anti-sigma-I factor RsgI | | Authors: | Brogan, A.P, Habib, C, Hobbs, S.J, Kranzusch, P.J, Rudner, D.Z. | | Deposit date: | 2023-06-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial SEAL domains undergo autoproteolysis and function in regulated intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6GZ8
 
 | | First GerMN domain of the sporulation protein GerM from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Spore germination protein GerM | | Authors: | Trouve, J, Mohamed, A, Leisico, F, Contreras-Martel, C, Liu, B, Mas, C, Rudner, D.Z, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural characterization of the sporulation protein GerM from Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6GZB
 
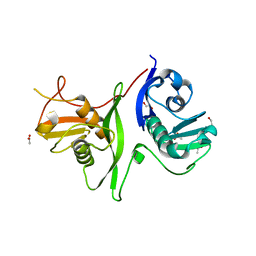 | | Tandem GerMN domains of the sporulation protein GerM from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Spore germination protein GerM | | Authors: | Trouve, J, Mohamed, A, Leisico, F, Contreras-Martel, C, Liu, B, Mas, C, Rudner, D.Z, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the sporulation protein GerM from Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6BAS
 
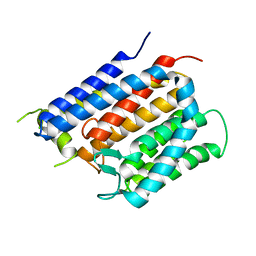 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | Descriptor: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
4C2E
 
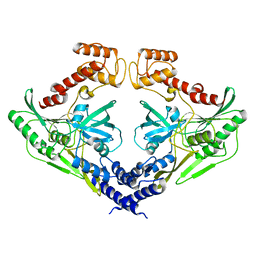 | | Crystal structure of the protease CtpB(S309A) present in a resting state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2C
 
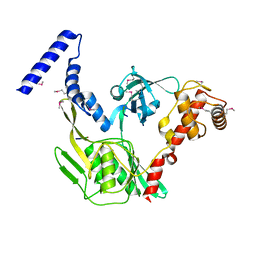 | | Crystal structure of the protease CtpB in an active state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2D
 
 | | Crystal structure of the protease CtpB in an active state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2G
 
 | | Crystal structure of CtpB(S309A) in complex with a peptide having a Val-Pro-Ala C-terminus | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2H
 
 | | Crystal structure of the CtpB(V118Y) mutant | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2F
 
 | | Crystal structure of the CtpB R168A mutant present in an active conformation | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
