1QRS
 
 | |
1QRU
 
 | |
2HAU
 
 | | Apo-Human Serum Transferrin (Non-Glycosylated) | | Descriptor: | CITRIC ACID, GLYCEROL, Serotransferrin | | Authors: | Wally, J, Everse, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding.
J.Biol.Chem., 281, 2006
|
|
2HAV
 
 | | Apo-Human Serum Transferrin (Glycosylated) | | Descriptor: | CITRIC ACID, GLYCEROL, Serotransferrin | | Authors: | Wally, J, Everse, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding.
J.Biol.Chem., 281, 2006
|
|
3RMA
 
 | | Crystal Structure of a replicative DNA polymerase bound to DNA containing Thymine Glycol | | Descriptor: | DNA (5'-D(*CP*GP*AP*(CTG)*GP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*CP*A)-3'), DNA polymerase | | Authors: | Aller, P, Duclos, S, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex.
J.Mol.Biol., 412, 2011
|
|
3RMC
 
 | | Crystal Structure of a replicative DNA polymerase bound to DNA containing Thymine Glycol | | Descriptor: | DNA (5'-D(*CP*GP*TP*(CTG)P*GP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*CP*A)-3'), DNA polymerase | | Authors: | Aller, P, Duclos, S, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex.
J.Mol.Biol., 412, 2011
|
|
3RMB
 
 | | Crystal Structure of a replicative DNA polymerase bound to DNA containing Thymine Glycol | | Descriptor: | DNA (5'-D(*CP*GP*CP*(CTG)P*GP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*CP*A)-3'), DNA polymerase, ... | | Authors: | Aller, P, Duclos, S, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex.
J.Mol.Biol., 412, 2011
|
|
1XMK
 
 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
1A1F
 
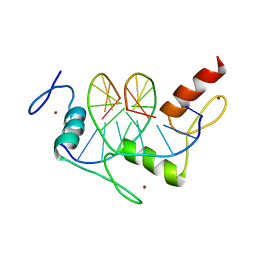 | | DSNR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*CP*CP*AP*CP*GP*C)-3'), THREE-FINGER ZIF268 PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1J
 
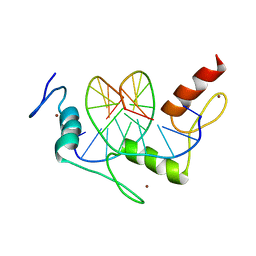 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (RADR ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1H
 
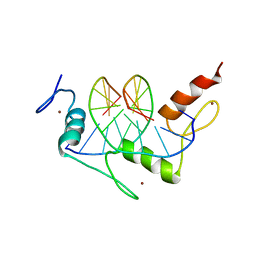 | | QGSR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), QGSR ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1G
 
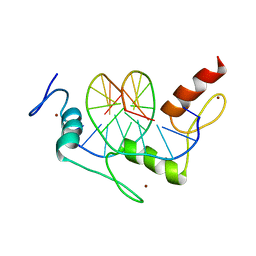 | | DSNR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), DSNR ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1I
 
 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), RADR ZIF268 ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1K
 
 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*CP*CP*AP*CP*GP*C)-3'), RADR ZIF268 VARIANT, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1L
 
 | | ZIF268 ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1BDV
 
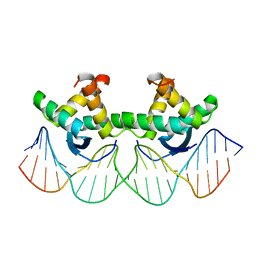 | | ARC FV10 COCRYSTAL | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (ARC FV10 REPRESSOR) | | Authors: | Schildbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1998-05-11 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CQT
 
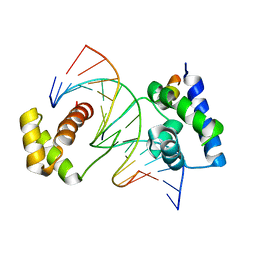 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX CONTAINING AN OCA-B PEPTIDE, THE OCT-1 POU DOMAIN, AND AN OCTAMER ELEMENT | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), POU DOMAIN, ... | | Authors: | Chasman, D.I, Cepek, K, Sharp, P.A, Pabo, C.O. | | Deposit date: | 1999-08-11 | | Release date: | 1999-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface.
Genes Dev., 13, 1999
|
|
1JK1
 
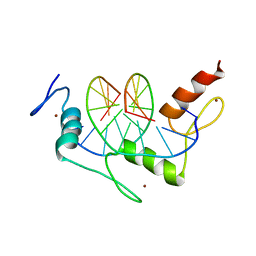 | | Zif268 D20A Mutant Bound to WT DNA Site | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*G)-3', 5'-D(*TP*CP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3', ZIF268, ... | | Authors: | Miller, J.C, Pabo, C.O. | | Deposit date: | 2001-07-11 | | Release date: | 2001-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition.
J.Mol.Biol., 313, 2001
|
|
1JK2
 
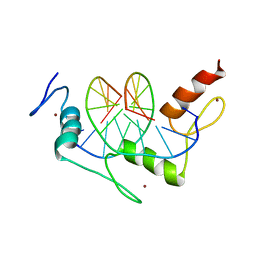 | | Zif268 D20A mutant bound to the GCT DNA site | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*CP*CP*CP*AP*CP*GP*C)-3', ZIF268, ... | | Authors: | Miller, J.C, Pabo, C.O. | | Deposit date: | 2001-07-11 | | Release date: | 2001-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition.
J.Mol.Biol., 313, 2001
|
|
1G2F
 
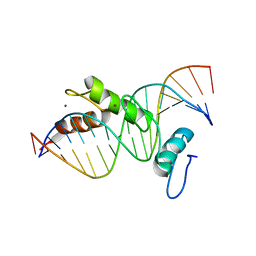 | | STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (TATAZF;CLONE #6) | | Descriptor: | 5'-D(*GP*AP*CP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*AP*G)-3', 5'-D(*TP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*GP*TP*CP*C)-3', TATA BOX ZINC FINGER PROTEIN, ... | | Authors: | Wolfe, S.A, Grant, R.A, Elrod-Erickson, M, Pabo, C.O. | | Deposit date: | 2000-10-18 | | Release date: | 2001-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Beyond the "recognition code": structures of two Cys2His2 zinc finger/TATA box complexes.
Structure, 9, 2001
|
|
1MYK
 
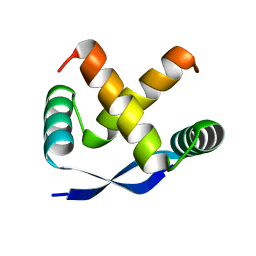 | | CRYSTAL STRUCTURE, FOLDING, AND OPERATOR BINDING OF THE HYPERSTABLE ARC REPRESSOR MUTANT PL8 | | Descriptor: | ARC REPRESSOR | | Authors: | Schildbach, J.F, Milla, M.E, Jeffrey, P.D, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1994-10-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure, folding, and operator binding of the hyperstable Arc repressor mutant PL8.
Biochemistry, 34, 1995
|
|
1MYL
 
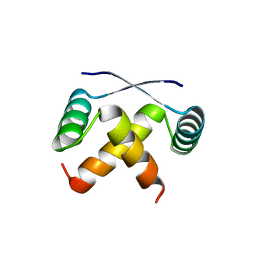 | |
1P47
 
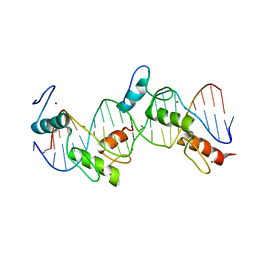 | | Crystal Structure of tandem Zif268 molecules complexed to DNA | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*CP*CP*AP*CP*GP*CP*CP*GP*CP*CP*CP*AP*CP*GP*CP*CP*A)-3', 5'-D(*GP*TP*GP*GP*CP*GP*TP*GP*GP*GP*CP*GP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3', Early growth response protein 1, ... | | Authors: | Peisach, E, Pabo, C.O. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Constraints for Zinc Finger Linker Design as Inferred from X-ray Crystal Structure of Tandem Zif268-DNA Complexes
J.Mol.Biol., 330, 2003
|
|
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
1P7J
 
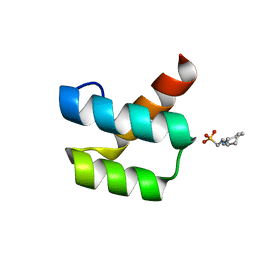 | | Crystal structure of engrailed homeodomain mutant K52E | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
