3IF9
 
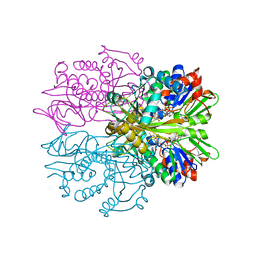 | | Crystal structure of Glycine Oxidase G51S/A54R/H244A mutant in complex with inhibitor glycolate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCOLIC ACID, Glycine oxidase | | Authors: | Pedotti, M, Rosini, E, Molla, G, Moschetti, T, Vallone, B, Savino, C, Pollegioni, L. | | Deposit date: | 2009-07-24 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glyphosate resistance by engineering the flavoenzyme glycine oxidase.
J.Biol.Chem., 284, 2009
|
|
4HST
 
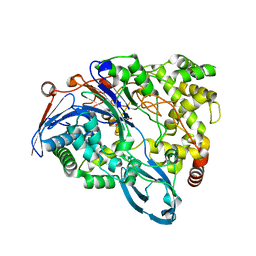 | | Crystal structure of a double mutant of a class III engineered cephalosporin acylase | | Descriptor: | 5,5-dihydroxy-L-norvaline, glutaryl-7-aminocephalosporanic acid acylase alpha chain, glutaryl-7-aminocephalosporanic acid acylase beta chain | | Authors: | Vrielink, A, Golden, E, Patterson, R, Tie, W.J, Anandan, A, Flematti, G, Molla, G, Rosini, E, Pollegioni, L. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Structure of a class III engineered cephalosporin acylase: comparisons with class I acylase and implications for differences in substrate specificity and catalytic activity.
Biochem.J., 451, 2013
|
|
4HSR
 
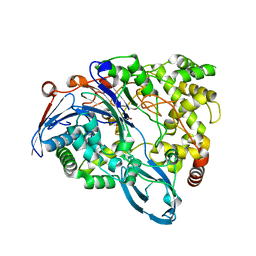 | | Crystal Structure of a class III engineered cephalosporin acylase | | Descriptor: | 5,5-dihydroxy-L-norvaline, glutaryl-7-aminocephalosporanic acid acylase alpha chain, glutaryl-7-aminocephalosporanic acid acylase beta chain | | Authors: | Vrielink, A, Golden, E, Patterson, R, Tie, W.J, Anandan, A, Flematti, G, Molla, G, Rosini, E, Pollegioni, L. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of a class III engineered cephalosporin acylase: comparisons with class I acylase and implications for differences in substrate specificity and catalytic activity.
Biochem.J., 451, 2013
|
|
6EGR
 
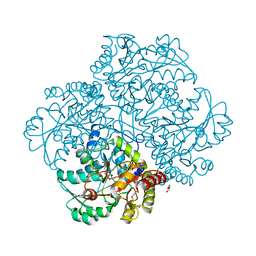 | | Crystal structure of Citrobacter freundii methionine gamma-lyase with V358Y replacement | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Revtovich, S.V, Demitri, N, Raboni, S, Nikulin, A.D, Morozova, E.A, Demidkina, T.V, Storici, P, Mozzarelli, A. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering methionine gamma-lyase from Citrobacter freundii for anticancer activity.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
