5K3V
 
 | | apo-PDX1.3 (Arabidopsis) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | Deposit date: | 2016-05-20 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2Z
 
 | | PDX1.3-adduct (Arabidopsis) | | Descriptor: | 1,2-ETHANEDIOL, 2-azanylpenta-1,4-dien-3-one, CHLORIDE ION, ... | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6HYE
 
 | | PDX1.2/PDX1.3 complex (PDX1.3:K97A) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6HXG
 
 | | PDX1.2/PDX1.3 complex (intermediate) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6HX3
 
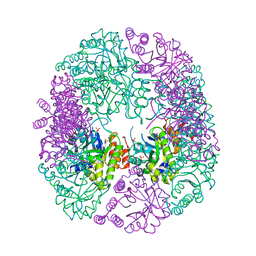 | | PDX1.2/PDX1.3 complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3ZIA
 
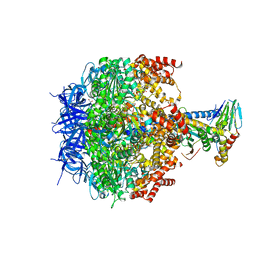 | | The structure of F1-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Robinson, G.C, Bason, J.V, Montgomery, M.G, Fearnley, I.M, Mueller, D.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2013-01-07 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of F1-ATPase from Saccharomyces Cerevisiae Inhibited by its Regulatory Protein If1.
Open Biol., 3, 2013
|
|
6EMK
 
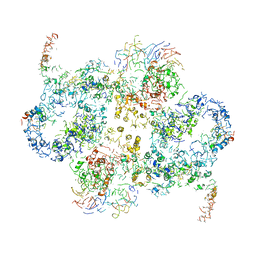 | | Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2 | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 2 subunit AVO1, Target of rapamycin complex 2 subunit AVO2, ... | | Authors: | Karuppasamy, M, Kusmider, B, Oliveira, T.M, Gaubitz, C, Prouteau, M, Loewith, R, Schaffitzel, C. | | Deposit date: | 2017-10-02 | | Release date: | 2017-12-06 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2.
Nat Commun, 8, 2017
|
|
