1OG3
 
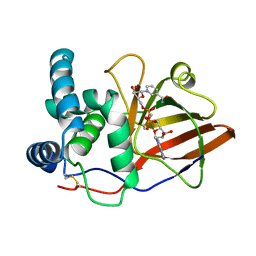 | | Crystal structure of the eukaryotic mono-ADP-ribosyltransferase ART2.2 mutant E189I in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, T-CELL ECTO-ADP-RIBOSYLTRANSFERASE 2 | | Authors: | Ritter, H, Koch-Nolte, F, Marquez, V.E, Schulz, G.E. | | Deposit date: | 2003-04-24 | | Release date: | 2003-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate Binding and Catalysis of Ecto-Adp-Ribosyltransferase 2.2 From Rat
Biochemistry, 42, 2003
|
|
1OG1
 
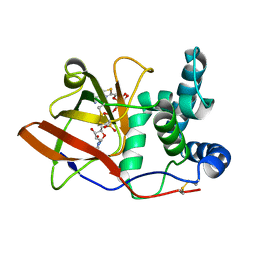 | | CRYSTAL STRUCTURE OF THE EUCARYOTIC MONO-ADP-RIBOSYLTRANSFERASE ART2.2 IN COMPLEX WITH TAD | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, T-CELL ECTO-ADP-RIBOSYLTRANSFERASE 2 | | Authors: | Ritter, H, Koch-Nolte, F, Marquez, V.E, Schulz, G.E. | | Deposit date: | 2003-04-23 | | Release date: | 2003-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Binding and Catalysis of Ecto-Adp-Ribosyltransferase 2.2 From Rat
Biochemistry, 42, 2003
|
|
1W27
 
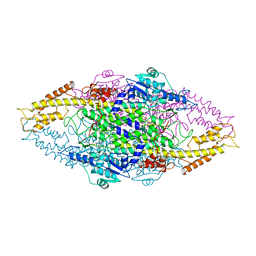 | | Phenylalanine ammonia-lyase (PAL) from Petroselinum crispum | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHENYLALANINE AMMONIA-LYASE 1 | | Authors: | Ritter, H, Schulz, G.E. | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Entrance Into the Phenylpropanoid Metabolism Catalyzed by Phenylalanine Ammonia-Lyase
Plant Cell, 16, 2004
|
|
1OG4
 
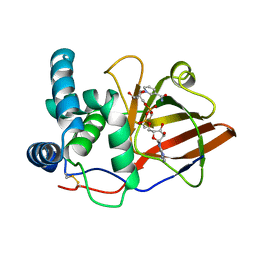 | | Crystal Structure of the Eucaryotic Mono-ADP-Ribosyltransferase ART2.2 Mutant E189A in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, T-CELL ECTO-ADP-RIBOSYLTRANSFERASE 2 | | Authors: | Ritter, H, Koch-Nolte, F, Marquez, V.E, Schulz, G.E. | | Deposit date: | 2003-04-24 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate Binding and Catalysis of Ecto-Adp-Ribosyltransferase 2.2 From Rat
Biochemistry, 42, 2003
|
|
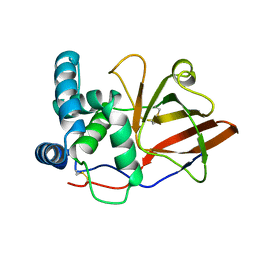
1GY0
 
 | |
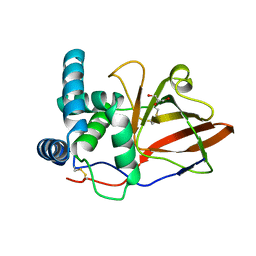
1GXY
 
 | |
1GXZ
 
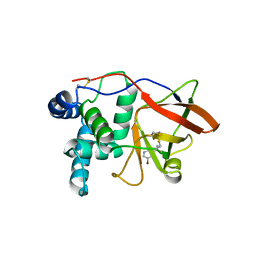 | |
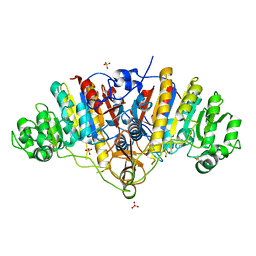
3BDG
 
 | |
3BDF
 
 | |
3BDH
 
 | |
