1EAX
 
 | | Crystal structure of MTSP1 (matriptase) | | Descriptor: | BENZAMIDINE, SULFATE ION, SUPPRESSOR OF TUMORIGENICITY 14 | | Authors: | Friedrich, R, Bode, W. | | Deposit date: | 2001-07-17 | | Release date: | 2002-01-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalytic Domain Structures of Mt-Sp1/Matriptase, a Matrix-Degrading Transmembrane Serine Proteinase.
J.Biol.Chem., 277, 2002
|
|
1NU9
 
 | | Staphylocoagulase-Prethrombin-2 complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
1NU7
 
 | | Staphylocoagulase-Thrombin Complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
1EAW
 
 | |
1EB1
 
 | | Complex structure of human thrombin with N-methyl-arginine inhibitor | | Descriptor: | 3-CYCLOHEXYL-D-ALANYL-L-PROLYL-N~2~-METHYL-L-ARGININE, PEPTIDE INHIBITOR, THROMBIN HEAVY CHAIN, ... | | Authors: | Friedrich, R, Steinmetzer, T, Bode, W. | | Deposit date: | 2001-07-18 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Methyl Group of N(Alpha)(Me)Arg-Containing Peptides Disturbs the Active-Site Geometry of Thrombin, Impairing Efficient Cleavage
J.Mol.Biol., 316, 2002
|
|
2A1D
 
 | | Staphylocoagulase bound to bovine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Friedrich, R, Panizzi, P, Kawabata, S, Bode, W, Bock, P.E, Fuentes-Prior, P. | | Deposit date: | 2005-06-20 | | Release date: | 2005-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis for Reduced Staphylocoagulase-mediated Bovine Prothrombin Activation
J.Biol.Chem., 281, 2006
|
|
1XM1
 
 | | Nonbasic Thrombin Inhibitor Complex | | Descriptor: | Hirudin, N-{[(2S)-1-(N-{[4-({[AMINO(IMINO)METHYL]AMINO}METHYL)CYCLOHEXYL]CARBONYL}-3-CYCLOHEXYL-L-ALANYL)AZETIDIN-2-YL]CARBONYL}-L-TYROSYL-N~6~-[AMINO(IMINO)METHYL]-L-LYSINAMIDE, thrombin | | Authors: | Friedrich, R, Bode, W, Schwienhorst, A. | | Deposit date: | 2004-10-01 | | Release date: | 2005-05-10 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nonbasic Thrombin Inhibitor Complex
To be Published
|
|
4COY
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Galb1-4GlcNAc | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4D3W
 
 | | Crystal Structure of Epithelial Adhesin 1 A domain (Epa1A) from Candida glabrata in complex with the T-antigen (Galb1-3GalNAc) | | Descriptor: | CALCIUM ION, EPITHELIAL ADHESIN 1, GLYCEROL, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-10-24 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4COU
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Lactose | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4CP2
 
 | | Crystal Structure of Epithelial Adhesin 9 A domain (Epa9A) from Candida glabrata in complex with Galb1-4GlcNAc | | Descriptor: | CALCIUM ION, CHLORIDE ION, EPITHELIAL ADHESIN 9, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional reprogramming of Candida glabrata epithelial adhesins: the role of conserved and variable structural motifs in ligand binding
To Be Published
|
|
4CP0
 
 | | Crystal Structure of Epithelial Adhesin 9 A domain (Epa9A) from Candida glabrata in complex with Lactose | | Descriptor: | CALCIUM ION, CHLORIDE ION, EPITHELIAL ADHESIN 9, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional reprogramming of Candida glabrata epithelial adhesins: the role of conserved and variable structural motifs in ligand binding
To Be Published
|
|
4COW
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with the T-antigen (Galb1-3GalNAc) | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4COZ
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Galb1-3GlcNAc | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4COV
 
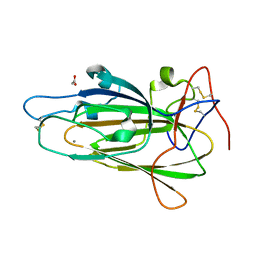 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Gala1-3Gal | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4CP1
 
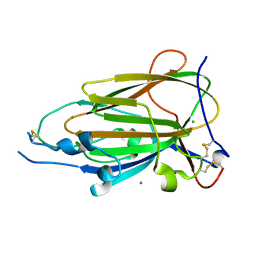 | | Crystal Structure of Epithelial Adhesin 9 A domain (Epa9A) from Candida glabrata in complex with Galb1-3GlcNAc | | Descriptor: | CALCIUM ION, CHLORIDE ION, EPITHELIAL ADHESIN 9, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional reprogramming of Candida glabrata epithelial adhesins: the role of conserved and variable structural motifs in ligand binding
To Be Published
|
|
1SAU
 
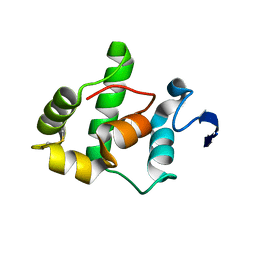 | | The Gamma subunit of the dissimilatory sulfite reductase (DsrC) from Archaeoglobus fulgidus at 1.1 A resolution | | Descriptor: | sulfite reductase, desulfoviridin-type subunit gamma | | Authors: | Mander, G.J, Weiss, M.S, Hedderich, R, Kahnt, J, Ermler, U, Warkentin, E. | | Deposit date: | 2004-02-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Determination of a novel structure by a combination of long-wavelength sulfur phasing and radiation-damage-induced phasing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4UZI
 
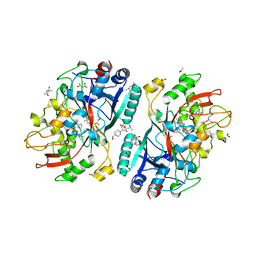 | | Crystal Structure of AauDyP Complexed with Imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Strittmatter, E, Liers, C, Ullrich, R, Hofrichter, M, Piontek, K, Plattner, D.A. | | Deposit date: | 2014-09-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Toolbox of Auricularia Auricula-Judae Dye-Decolorizing Peroxidase - Identification of Three New Potential Substrate-Interaction Sites.
Arch.Biochem.Biophys., 574, 2015
|
|
4BEB
 
 | | MUTANT (K220E) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
6Y98
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 9 to 1 A domain (Epa9-CBL2Epa1) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, PA14 domain-containing protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
6Y9J
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 9 A domain (Epa1-CBL2Epa9) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epa1p, ... | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
4YQE
 
 | | Crystal structure of E. coli WrbA in complex with benzoquinone | | Descriptor: | 1,4-benzoquinone, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | Authors: | Degtjarik, O, Brynda, J, Ettrichova, O, Carey, J, Kuta Smatanova, I, Ettrich, R. | | Deposit date: | 2015-03-13 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Quantum Calculations Indicate Effective Electron Transfer between FMN and Benzoquinone in a New Crystal Structure of Escherichia coli WrbA.
J.Phys.Chem.B, 120, 2016
|
|
7A46
 
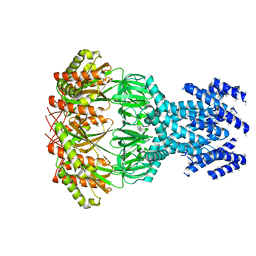 | | small conductance mechanosensitive channel YbiO | | Descriptor: | Putative transport protein | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZYD
 
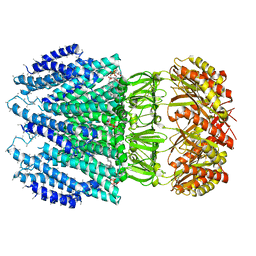 | | YnaI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI,Low conductance mechanosensitive channel YnaI | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-07-31 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZYE
 
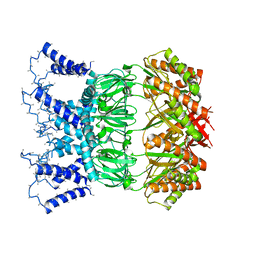 | | YnaI in an open-like conformation | | Descriptor: | YnaI,Low conductance mechanosensitive channel YnaI | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-07-31 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
