3KSO
 
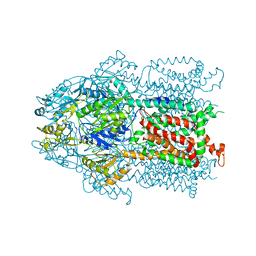 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | Cation efflux system protein cusA, SILVER ION | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.367 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
3KSS
 
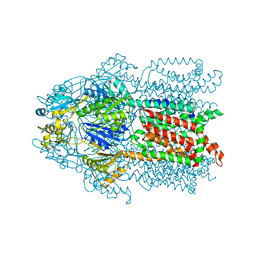 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | COPPER (I) ION, Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
3KYH
 
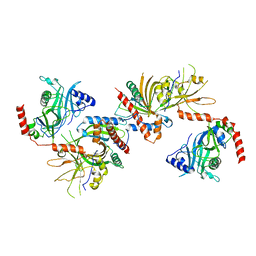 | | Saccharomyces cerevisiae Cet1-Ceg1 capping apparatus | | Descriptor: | mRNA-capping enzyme subunit alpha, mRNA-capping enzyme subunit beta | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA Capping Apparatus.
Structure, 18, 2010
|
|
5UI8
 
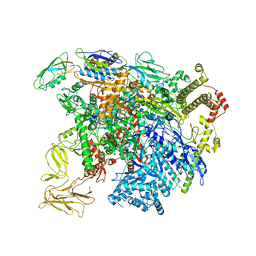 | | structure of sigmaN-holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Crystal structure of Aquifex aeolicus sigma (N) bound to promoter DNA and the structure of sigma (N)-holoenzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4R1I
 
 | |
3MBW
 
 | | Crystal structure of the human ephrin A2 LBD and CRD domains in complex with ephrin A1 | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1, UNKNOWN ATOM OR ION, ... | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4R0C
 
 | | Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology | | Descriptor: | AbgT putative transporter family, DODECYL-BETA-D-MALTOSIDE, SODIUM ION, ... | | Authors: | Su, C.-C, Bolla, J.R, Yu, E.W. | | Deposit date: | 2014-07-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology.
Nat Commun, 6, 2015
|
|
5BSA
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.611 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5BS7
 
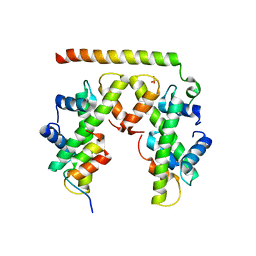 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
6CAM
 
 | |
6BMM
 
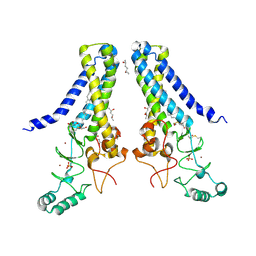 | | Structure of human DHHC20 palmitoyltransferase, space group P21 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,5S)-hexane-2,5-diol, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
1X94
 
 | |
4DOH
 
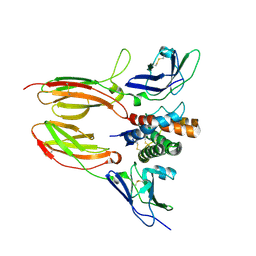 | | IL20/IL201/IL20R2 Ternary Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-20, Interleukin-20 receptor subunit alpha, ... | | Authors: | Logsdon, N.J, Walter, M.R. | | Deposit date: | 2012-02-09 | | Release date: | 2012-07-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for receptor sharing and activation by interleukin-20 receptor-2 (IL-20R2) binding cytokines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DOP
 
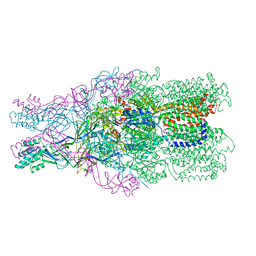 | | Crystal structure of the CusBA heavy-metal efflux complex from Escherichia coli, R mutant | | Descriptor: | Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E. | | Deposit date: | 2012-02-10 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
4DNT
 
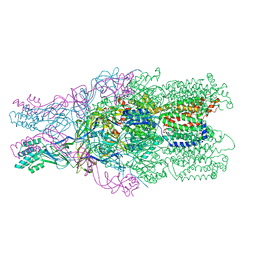 | | Crystal structure of the CusBA heavy-metal efflux complex from Escherichia coli, mutant | | Descriptor: | Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E. | | Deposit date: | 2012-02-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
3PJZ
 
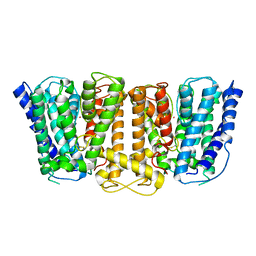 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
4MT0
 
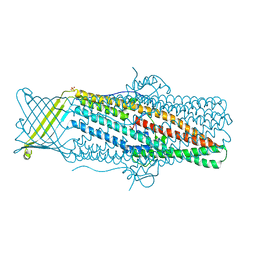 | |
5D1R
 
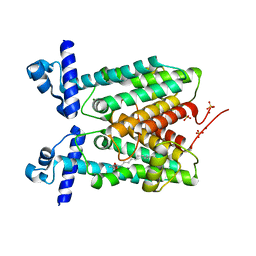 | | Crystal structure of Mycobacterium tuberculosis Rv1816 transcriptional regulator. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Rv1816 transcriptional regulator, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
3OPO
 
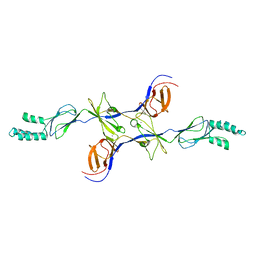 | |
3NE5
 
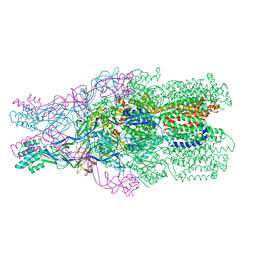 | |
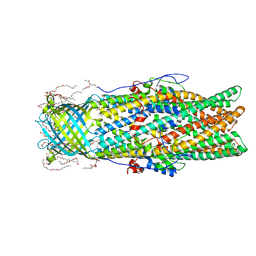
4MT4
 
 | |
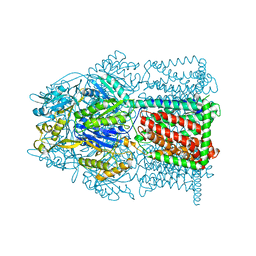
4MT1
 
 | |
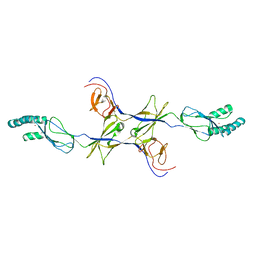
3OW7
 
 | |
5D18
 
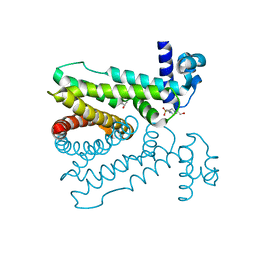 | | Crystal structure of Mycobacterium tuberculosis Rv0302, form I | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis transcriptional regulator Rv0302.
Protein Sci., 2015
|
|
3Q9E
 
 | | Crystal structure of H159A APAH complexed with acetylspermine | | Descriptor: | Acetylpolyamine amidohydrolase, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, POTASSIUM ION, ... | | Authors: | Lombardi, P.M, Christianson, D.W. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases .
Biochemistry, 50, 2011
|
|
