1MMI
 
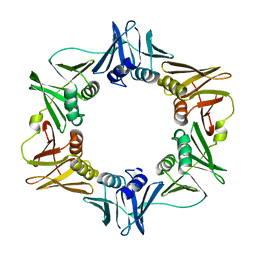 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3QSB
 
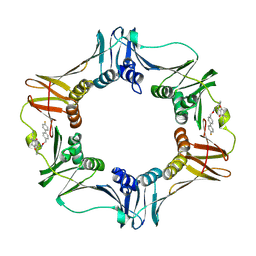 | | Structure of E. coli polIIIbeta with (Z)-5-(1-((4'-Fluorobiphenyl-4-yl)methoxyimino)butyl)-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile | | Descriptor: | (1R,5R)-5-{(1Z)-N-[(4'-fluorobiphenyl-4-yl)methoxy]butanimidoyl}-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile, DNA polymerase III subunit beta | | Authors: | Wijffels, G, Johnson, W.M, Oakley, A.J, Turner, K, Epa, V.C, Briscoe, S.J, Polley, M, Liepa, A.J, Hofmann, A, Buchardt, J, Christensen, C, Prosselkov, P, Dalrymple, B.P, Alewood, P.F, Jennings, P.A, Dixon, N.E, Winkler, D.A. | | Deposit date: | 2011-02-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding inhibitors of the bacterial sliding clamp by design
J.Med.Chem., 54, 2011
|
|
1T3W
 
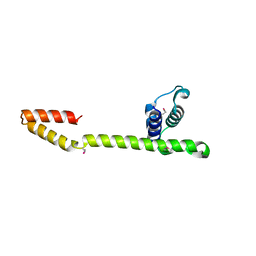 | | Crystal Structure of the E.coli DnaG C-terminal domain (residues 434 to 581) | | Descriptor: | ACETIC ACID, DNA primase | | Authors: | Oakley, A.J, Loscha, K.V, Schaeffer, P.M, Liepinsh, E, Wilce, M.C.J, Otting, G, Dixon, N.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal and solution structures of the helicase-binding domain of Escherichia coli primase
J.Biol.Chem., 280, 2005
|
|
