4HEB
 
 | | The Crystal structure of Maf protein of Bacillus subtilis | | Descriptor: | Septum formation protein Maf, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dombrovski, L, Brown, G, Flick, R, Tchigvintsev, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Iakounine, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
4IC1
 
 | | Crystal structure of SSO0001 | | Descriptor: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-09 | | Release date: | 2013-01-16 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
4Z8X
 
 | | Truncated FtsH from A. aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, SULFATE ION, ... | | Authors: | Vostrukhina, M, Baumann, U, Schacherl, M, Bieniossek, C, Lanz, M, Baumgartner, R. | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The structure of Aquifex aeolicus FtsH in the ADP-bound state reveals a C2-symmetric hexamer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5M00
 
 | | Crystal structure of murine P14 TCR complex with H-2Db and Y4A, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
5M01
 
 | | Crystal structure of murine P14 TCR/ H-2Db complex with PA, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
5M02
 
 | |
4PXK
 
 | | Crystal structure of Haloarcula marismortui bacteriorhodopsin I D94N mutant | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Shevchenko, V, Gushchin, I, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2014-03-24 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Escherichia coli-Expressed Haloarcula marismortui Bacteriorhodopsin I in the Trimeric Form.
Plos One, 9, 2014
|
|
6RF5
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric form, pH 6.0 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF6
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric form, pH 8.0 | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RFA
 
 | | Crystal structure of the H30K mutant of the light-driven sodium pump KR2 in the monomeric form, pH 8.0 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6REZ
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form, pH 5.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF7
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric form, pH 8.9 | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RFB
 
 | | Crystal structure of the potassium-pumping S254A mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.3 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF1
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric "wet" form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF9
 
 | | Crystal structure of the Y154F mutant of the light-driven sodium pump KR2 in the monomeric form, pH 8.0 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RFC
 
 | | Crystal structure of the potassium-pumping G263F mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF0
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric "dry" form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6REX
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form, pH 6.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF4
 
 | | Crystal structure of the potassium-pumping S254A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF3
 
 | | Crystal structure of the potassium-pumping G263F mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, SODIUM ION, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
4E48
 
 | |
6WGD
 
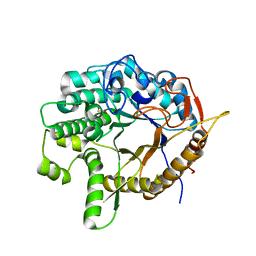 | | Crystal structure of a 6-phospho-beta-glucosidase from Bacillus licheniformis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2020-04-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structure, Bioinformatics Analysis, and Substrate Specificity of a 6-Phospho-beta-glucosidase Glycoside Hydrolase 1 Enzyme from Bacillus licheniformis .
J.Chem.Inf.Model., 60, 2020
|
|
7M1R
 
 | |
1SH6
 
 | |
1SH5
 
 | |
