4LVJ
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt). Mn-bound crystal structure at pH 5.5 | | Descriptor: | ACETATE ION, ACTTTAT oligonucleotide, ATAAAGTATAGTGTG oligonucleotide, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVI
 
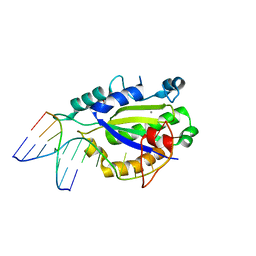 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTG oligonucleotide, GLYCEROL, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVL
 
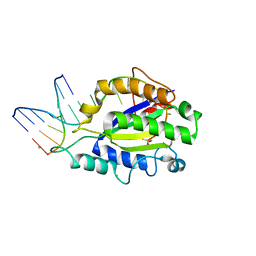 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Thiophosphate). Mn-bound crystal structure at pH 6.8 | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*CP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*GP*TP*AP*TP*AP*GP*TP*GP*TP*GP*(TS6))-3'), ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6RC9
 
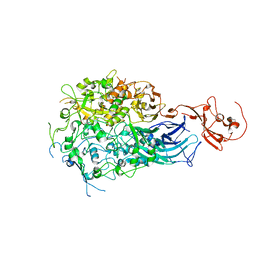 | |
6RJ1
 
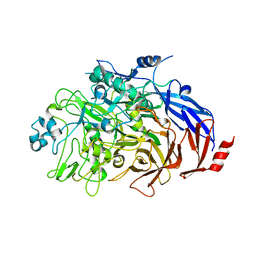 | |
4HHH
 
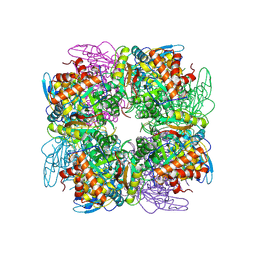 | | Structure of Pisum sativum Rubisco | | Descriptor: | RIBULOSE-1,5-DIPHOSPHATE, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Loewen, P.C, Didychuk, A.L, Switala, J, Loewen, M.C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-10-31 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Pisum sativum Rubisco with bound ribulose 1,5-bisphosphate.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2QLW
 
 | |
2QLX
 
 | |
6RCC
 
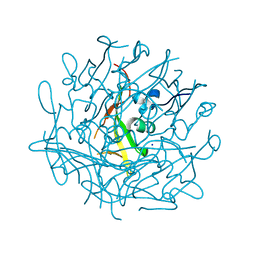 | | Domain C P140 Mycoplasma genitalium | | Descriptor: | Adhesin P1, CHLORIDE ION, SODIUM ION | | Authors: | Vizarraga, D, Aparicio, D, Perez, R, Illanes, R, Fita, I. | | Deposit date: | 2019-04-11 | | Release date: | 2020-11-04 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Alternative conformation of the C-domain of the P140 protein from Mycoplasma genitalium.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6RCD
 
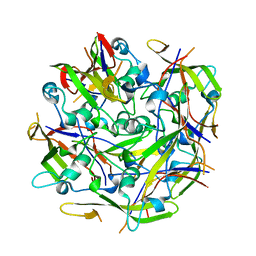 | | Octamer C-Domain P140 Mycoplasma genitalium. | | Descriptor: | MgPa adhesin | | Authors: | Vizarraga, D, Aparicio, D, Perez, R, Illanes, R, Fita, I. | | Deposit date: | 2019-04-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Alternative conformation of the C-domain of the P140 protein from Mycoplasma genitalium.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1ZM5
 
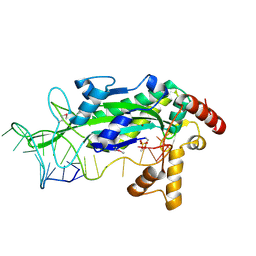 | | Conjugative Relaxase TRWC in complex with ORIT dna, cooper-bound structure | | Descriptor: | COPPER (II) ION, DNA (25-MER), SULFATE ION, ... | | Authors: | Boer, R, Russi, S, Guasch, A, Lucas, M, Blanco, A.G, Coll, M, de la Cruz, F. | | Deposit date: | 2005-05-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unveiling the Molecular Mechanism of a Conjugative Relaxase: The Structure of TrwC Complexed with a 27-mer DNA Comprising the Recognition Hairpin and the Cleavage Site
J.Mol.Biol., 358, 2006
|
|
2WYL
 
 | | Apo structure of a metallo-b-lactamase | | Descriptor: | FORMYL GROUP, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
2WYM
 
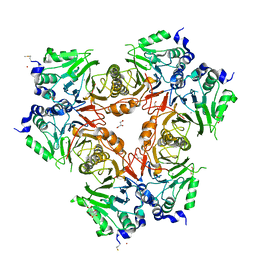 | | Structure of a metallo-b-lactamase | | Descriptor: | CITRATE ANION, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG, ... | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
2BI4
 
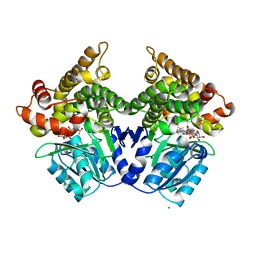 | | Lactaldehyde:1,2-propanediol oxidoreductase of Escherichia coli | | Descriptor: | CHLORIDE ION, FE (III) ION, LACTALDEHYDE REDUCTASE, ... | | Authors: | Montella, C, Bellsolell, L, Badia, J, Baldoma, L, Perez, R, Coll, M, Aguilar, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of an Iron-Dependent Group III Dehydrogenase that Interconverts L-Lactaldehyde and L-1,2-Propanediol in Escherichia Coli
J.Bacteriol., 187, 2005
|
|
1E9R
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Trigonal form in complex with sulphate. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB, SULFATE ION | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1E9S
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Unbound monoclinic form. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GL6
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolysable GTP analogue GDPNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATPASE, CHLORIDE ION, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GL7
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolisable ATP-analogue ADPNP. | | Descriptor: | CHLORIDE ION, CONJUGAL TRANSFER PROTEIN TRWB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GKI
 
 | | Plasmid coupling protein TrwB in complex with ADP and Mg2+. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, CONJUGAL TRANSFER PROTEIN TRWB, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
6R21
 
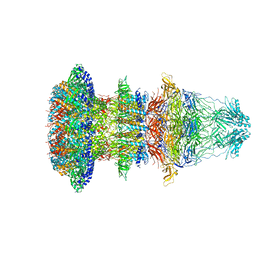 | | Cryo-EM structure of T7 bacteriophage fiberless tail complex | | Descriptor: | Portal protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Cuervo, A, Fabrega-Ferrer, M, Machon, C, Conesa, J.J, Perez-Ruiz, M, Coll, M, Carrascosa, J.L. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
1EA4
 
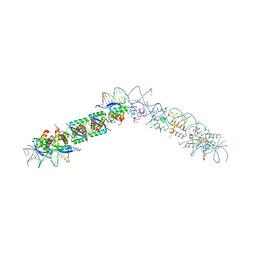 | | TRANSCRIPTIONAL REPRESSOR COPG/22bp dsDNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*GP*TP*GP *CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*TP*C)-3'), DNA(5'-D(*AP*GP*AP*TP*TP*GP*CP*AP*TP *TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*GP*TP*T)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Costa, M, Sola, M, Acebo, P, Eritja, R, Espinosa, M, Solar, G.D, Coll, M. | | Deposit date: | 2000-11-05 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasmid Transcriptional Repressor Copg Oligomerises to Render Helical Superstructures Unbound and in Complexes with Oligonucleotides
J.Mol.Biol., 310, 2001
|
|
2CPG
 
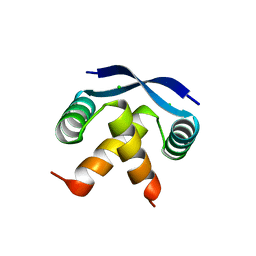 | | TRANSCRIPTIONAL REPRESSOR COPG | | Descriptor: | CHLORIDE ION, TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Sola, M, Acebo, P, Parraga, A, Guasch, A, Eritja, R, Gonzalez, A, Espinosa, M, del Solar, G, Coll, M. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator.
EMBO J., 17, 1998
|
|
