7QAN
 
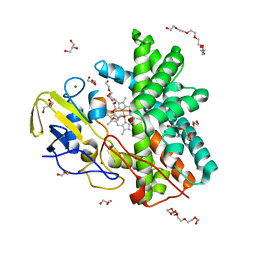 | | Cytochrome P450 Enzyme AbyV | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Parnell, A.E, Back, C.R, Race, P.R. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Role of Cytochrome P450 AbyV in the Final Stages of Abyssomicin C Biosynthesis.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4YEG
 
 | |
5LLF
 
 | |
5LLB
 
 | |
5LL0
 
 | |
7AH0
 
 | | Crystal structure of the de novo designed two-heme binding protein, 4D2 | | Descriptor: | 4D2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hutchins, G.H, Parnell, A.E, Anderson, J.L.R. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expandable, modular de novo protein platform for precision redox engineering.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6YZG
 
 | |
5LC9
 
 | |
5LDB
 
 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5LCD
 
 | | Structure of Polyphosphate Kinase from Meiothermus ruber bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-06-21 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5LD1
 
 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-06-23 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MAQ
 
 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ADP and PPi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5O6K
 
 | |
5O6M
 
 | |
8CCR
 
 | |
6SZC
 
 | |
4YWF
 
 | | AbyA5 | | Descriptor: | AbyA5 | | Authors: | Byrne, M.J, Race, P.R. | | Deposit date: | 2015-03-20 | | Release date: | 2016-06-29 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Esterase-like Lyase Catalyzes Acetate Elimination in Spirotetronate/Spirotetramate Biosynthesis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5NO5
 
 | | AbyA5 Wildtype | | Descriptor: | AbyA5 | | Authors: | Byrne, M.J, Race, P.R. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Esterase-like Lyase Catalyzes Acetate Elimination in Spirotetronate/Spirotetramate Biosynthesis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
