6R5J
 
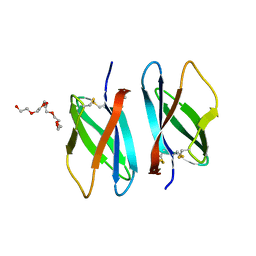 | |
2JOB
 
 | | Solution structure of an antilipopolysaccharide factor from shrimp and its possible Lipid A binding site | | Descriptor: | antilipopolysaccharide factor | | Authors: | Yang, Y, Boze, H, Chemardin, P, Padilla, A, Moulin, G, Tassanakajon, A, Pugniere, M, Roquet, F, Gueguen, Y, Bachere, E, Aumelas, A. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: Model of the possible lipid A-binding site
Biopolymers, 91, 2009
|
|
1R3B
 
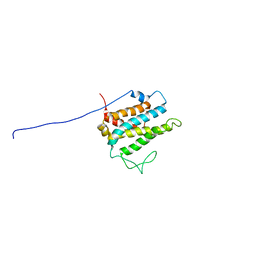 | | Solution structure of xenopus laevis Mob1 | | Descriptor: | MOB1 | | Authors: | Ponchon, L, Dumas, C, Kajava, A.V, Fesquet, D, Padilla, A. | | Deposit date: | 2003-10-01 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Mob1, a mitotic exit network protein and its interaction with an NDR kinase peptide
J.Mol.Biol., 337, 2004
|
|
1T6F
 
 | | Crystal Structure of the Coiled-coil Dimerization Motif of Geminin | | Descriptor: | Geminin | | Authors: | Thepaut, M, Maiorano, D, Guichou, J.-F, Auge, M.-T, Dumas, C, Mechali, M, Padilla, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of the Coiled-coil Dimerization Motif of Geminin: Structural and Functional Insights on DNA Replication Regulation
J.Mol.Biol., 342, 2004
|
|
7L7A
 
 | | Solution Structure of NuxVA | | Descriptor: | NuxVA | | Authors: | Mari, F, Dovell, S. | | Deposit date: | 2020-12-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conus venom fractions inhibit the adhesion of Plasmodium falciparum erythrocyte membrane protein 1 domains to the host vascular receptors.
J Proteomics, 234, 2020
|
|
1PVA
 
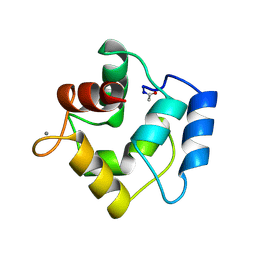 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN (ALPHA COMPONENT FROM PIKE MUSCLE) | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Roquet, F, Rambaud, J, Parello, J. | | Deposit date: | 1995-01-16 | | Release date: | 1995-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin (Alpha Component from Pike Muscle)
To be Published
|
|
5ZNG
 
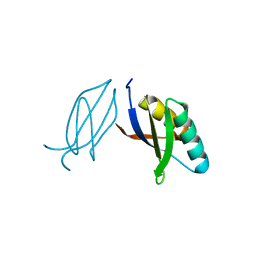 | | The crystal complex of immune receptor RGA5A_S of Pia from rice (Oryzae sativa) with rice blast (Magnaporthe oryzae) effector protein AVR1-CO39 | | Descriptor: | AVR1-CO39, NBS-LRR type protein | | Authors: | Guo, L.W, Zhang, Y.K, Liu, Q, Ma, M.Q, Liu, J.F, Peng, Y.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZNE
 
 | |
4QFT
 
 | |
6H0I
 
 | | Solution structure of Melampsora larici-populina MlpP4.1 | | Descriptor: | Secreted protein | | Authors: | Tsan, P, Petre, B, Hecker, A, Rouhier, N, Duplessis, S. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds.
Sci Rep, 9, 2019
|
|
2MYW
 
 | |
2MYV
 
 | |
4JMI
 
 | |
4JWL
 
 | |
4JMO
 
 | |
4JXH
 
 | |
4L5M
 
 | |
2LC0
 
 | | Rv0020c_Nter structure | | Descriptor: | Putative uncharacterized protein TB39.8 | | Authors: | Barthe, P.P, Cohen-Gonsaud, M.M, Roumestand, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-11-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Mycobacterium tuberculosis Rv0020c Protein and Its Interaction with the PknB Kinase
Structure, 19, 2011
|
|
2LC1
 
 | | Rv0020c_FHA Structure | | Descriptor: | Putative uncharacterized protein TB39.8 | | Authors: | Barthe, P.P, Cohen-Gonsaud, M.M, Roumestand, C.C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-11-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Mycobacterium tuberculosis Rv0020c Protein and Its Interaction with the PknB Kinase
Structure, 19, 2011
|
|
