5V68
 
 | | Crystal structure of cell division protein FtsZ from Mycobacterium tuberculosis bounded via the T9 loop | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Lazo, E.O, Ojima, I, Chowdhury, S.R, Awasthi, D, Jakoncic, J. | | Deposit date: | 2017-03-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Novel T9 loop conformation of filamenting temperature-sensitive mutant Z from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4AZN
 
 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag-mediated crystal packing | | Descriptor: | FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZO
 
 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag removed | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZQ
 
 | | Murine epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid 2-arachidonoylglycerol | | Descriptor: | 2-hydroxy-1-(hydroxymethyl)ethyl icosanoate, CHLORIDE ION, FATTY ACID-BINDING PROTEIN, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZM
 
 | | Human epidermal fatty acid-binding protein (FABP5) in complex with the inhibitor BMS-309413 | | Descriptor: | ((2'-(5-ETHYL-3,4-DIPHENYL-1H-PYRAZOL-1-YL)-3-BIPHENYLYL)OXY)ACETIC ACID, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZP
 
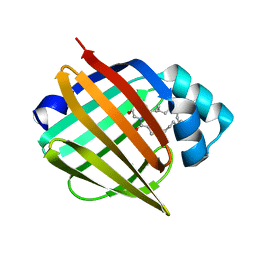 | | Murine epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid anandamide | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZR
 
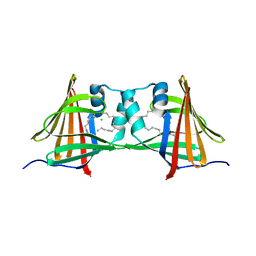 | | Human epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid anandamide | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5UR9
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP5 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, epidermal, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19800353 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
8EXD
 
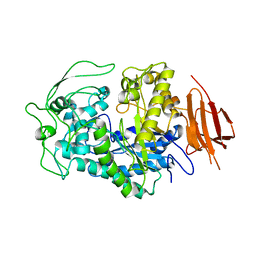 | |
6AQ1
 
 | | The crystal structure of human FABP3 | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2017-08-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.40000093 Å) | | Cite: | SAR studies on truxillic acid mono esters as a new class of antinociceptive agents targeting fatty acid binding proteins.
Eur J Med Chem, 154, 2018
|
|
5URA
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP7 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, brain, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85002172 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
7LPQ
 
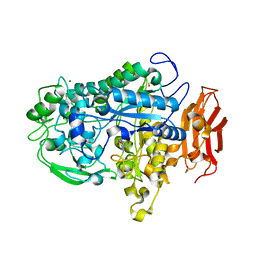 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with hit 9 | | Descriptor: | Cytoplasmic protein, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
7LPO
 
 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytoplasmic protein, MAGNESIUM ION | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
7LPP
 
 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with hit 1 | | Descriptor: | 4-(hydroxymethyl)-1-[[2-(3-methoxyphenyl)-1,3-thiazol-5-yl]methyl]piperidin-4-ol, Cytoplasmic protein, GLYCEROL, ... | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
2Q1Y
 
 | |
2Q1X
 
 | |
8WMD
 
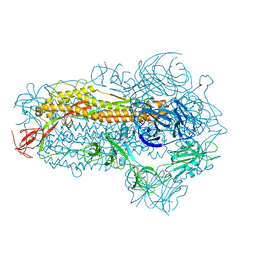 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
8WMF
 
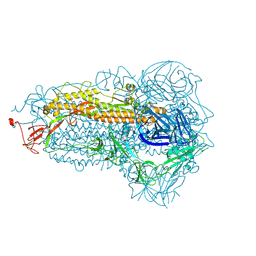 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
8XLM
 
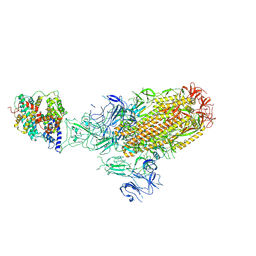 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-12-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
8XLN
 
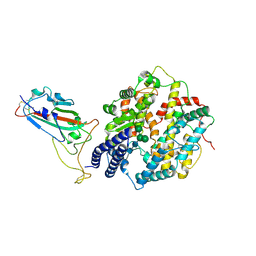 | | Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-12-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
