7C51
 
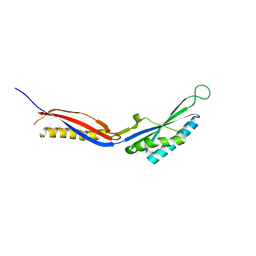 | |
7C50
 
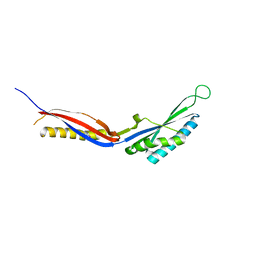 | |
7X1K
 
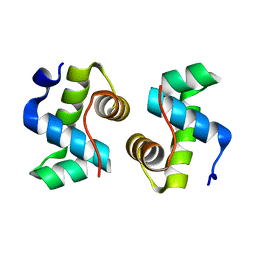 | |
7W1F
 
 | | Crystal structure of the dNTP triphosphohydrolase PA1124 from Pseudomonas aeruginosa | | Descriptor: | NICKEL (II) ION, Probable deoxyguanosinetriphosphate triphosphohydrolase | | Authors: | Oh, H.B, Song, W.S, Lee, K.C, Park, S.C, Yoon, S.I. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the dNTP triphosphohydrolase PA1124 from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7X9R
 
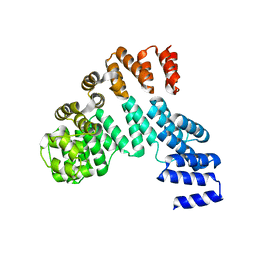 | | Crystal structure of the antirepressor GmaR | | Descriptor: | Glycosyl transferase family 2 | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7X9S
 
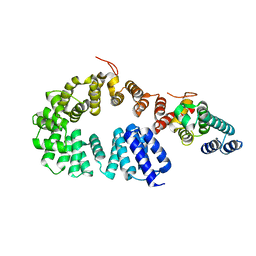 | | Crystal structure of a complex between the antirepressor GmaR and the transcriptional repressor MogR | | Descriptor: | GmaR, Motility gene repressor MogR | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
8J56
 
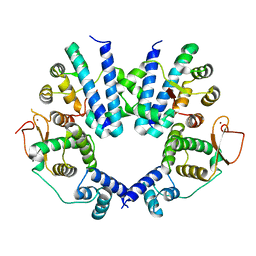 | | Crystal structure of the FlhDC complex from Cupriavidus necator | | Descriptor: | Flagellar transcriptional regulator FlhC, Flagellar transcriptional regulator FlhD, ZINC ION | | Authors: | Cho, S.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-04-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hexameric structure of the flagellar master regulator FlhDC from Cupriavidus necator and its interaction with flagellar promoter DNA.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
7YMO
 
 | |
8JZC
 
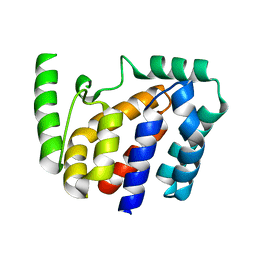 | | Crystal structure of Geobacillus stearothermophilus NarJ | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
8JZD
 
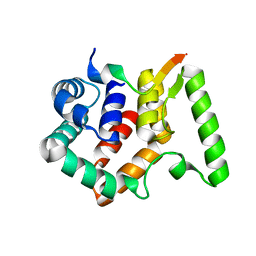 | | Crystal structure of Escherichia coli NarJ in complex with the signal peptide of E. coli NarG | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone NarJ, Respiratory nitrate reductase 1 alpha chain | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
4EDA
 
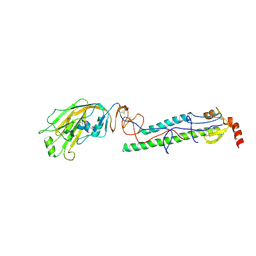 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
4EDB
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
4F15
 
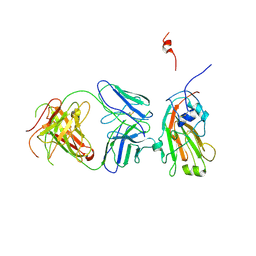 | | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-05-06 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses
To be Published
|
|
8K3F
 
 | |
4KIA
 
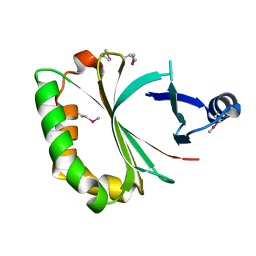 | | Crystal structure of LmHde, heme-degrading enzyme, from Listeria monocytogenes | | Descriptor: | Lmo2213 protein | | Authors: | Kim, K.K, Duong, T, Kim, T. | | Deposit date: | 2013-05-02 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional characterization of an Isd-type haem-degradation enzyme from Listeria monocytogenes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6IWY
 
 | |
6J3D
 
 | |
