3AUH
 
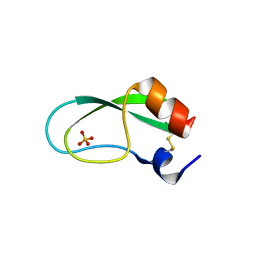 | | A simplified BPTI variant with poly Arg amino acid tag (C3R) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUE
 
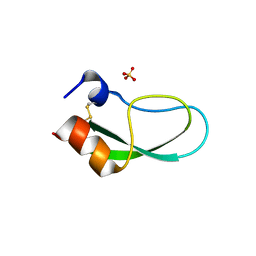 | | A simplified BPTI variant with poly His amino acid tag (C5H) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AEI
 
 | | Crystal structure of the prefoldin beta2 subunit from Thermococcus strain KS-1 | | Descriptor: | CHLORIDE ION, Prefoldin beta subunit 2, SULFATE ION | | Authors: | Ohtaki, A, Sugano, Y, Sato, T, Noguchi, K, Miyatake, H, Yohda, M. | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic Characterization of the Interaction between Prefoldin and Group II Chaperonin
J.Mol.Biol., 399, 2010
|
|
3X20
 
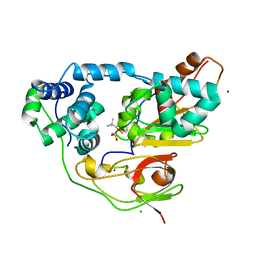 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 25 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, N, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6J1A
 
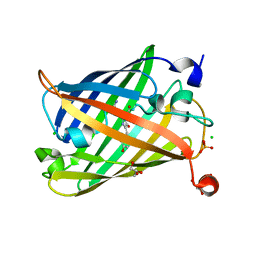 | | Photoswitchable fluorescent protein Gamillus, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6JXF
 
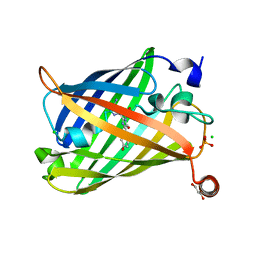 | | Photoswitchable fluorescent protein Gamillus, off-state (pH7.0) | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1C
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1B
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, on-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
2DRX
 
 | | Structure Analysis of (POG)4-(LOG)2-(POG)4 | | Descriptor: | collagen like peptide | | Authors: | Okuyama, K. | | Deposit date: | 2006-06-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unique side chain conformation of a leu residue in a triple-helical structure
Biopolymers, 86, 2007
|
|
2DRT
 
 | | Structure Analysis of (POG)4-LOG-(POG)5 | | Descriptor: | ETHANOL, collagen like peptide | | Authors: | Okuyama, K. | | Deposit date: | 2006-06-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unique side chain conformation of a leu residue in a triple-helical structure
Biopolymers, 86, 2007
|
|
3A19
 
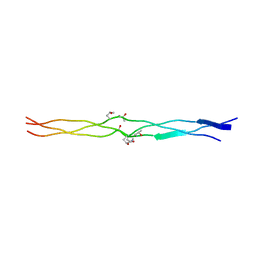 | | Structure of (PPG)4-OOG-(PPG)4_H monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hyakutake, M, Wu, G, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-03-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two crystal modifications of (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 reveal the puckering preference of Hyp(X) in the Hyp(X):Hyp(Y) and Hyp(X):Pro(Y) stacking pairs in collagen helices.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2ZDI
 
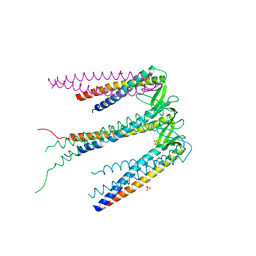 | | Crystal structure of Prefoldin from Pyrococcus horikoshii OT3 | | Descriptor: | Prefoldin subunit alpha, Prefoldin subunit beta, SULFATE ION | | Authors: | Kida, H, Miki, K. | | Deposit date: | 2007-11-23 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and molecular dynamics simulation of archaeal prefoldin: the molecular mechanism for binding and recognition of nonnative substrate proteins
J.Mol.Biol., 376, 2008
|
|
3APT
 
 | |
3APY
 
 | |
3VTT
 
 | |
3A3G
 
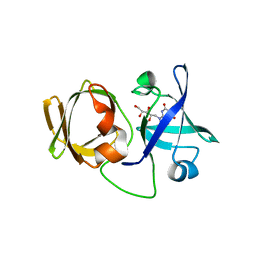 | | Crystal structure of LumP complexed with 6,7-dimethyl-8-(1'-D-ribityl) lumazine | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Lumazine protein | | Authors: | Sato, Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3A3B
 
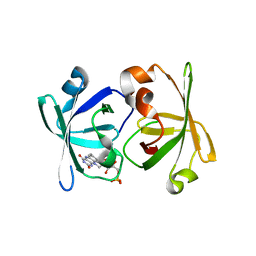 | | Crystal structure of LumP complexed with flavin mononucleotide | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lumazine protein, RIBOFLAVIN | | Authors: | Sato, Y. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3A35
 
 | | Crystal structure of LumP complexed with riboflavin | | Descriptor: | Lumazine protein, RIBOFLAVIN | | Authors: | Sato, Y. | | Deposit date: | 2009-06-09 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
