3KDO
 
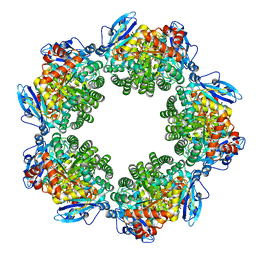 | | Crystal structure of Type III Rubisco SP6 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
3KDN
 
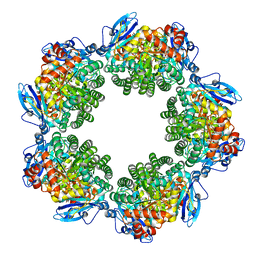 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
4GA6
 
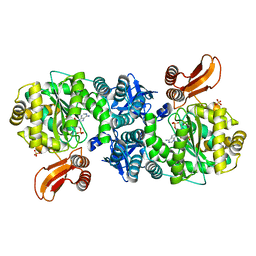 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in complex with substrates | | Descriptor: | ADENOSINE MONOPHOSPHATE, Putative thymidine phosphorylase, SULFATE ION | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
4GA4
 
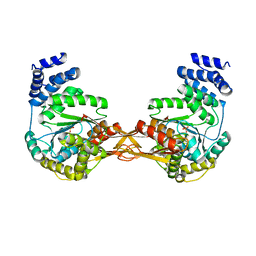 | | Crystal structure of AMP phosphorylase N-terminal deletion mutant | | Descriptor: | PHOSPHATE ION, Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
5HEE
 
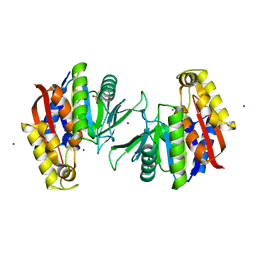 | | Crystal structure of the TK2203 protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, TK2203 protein, ... | | Authors: | Nishitani, Y, Miki, K. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4GA5
 
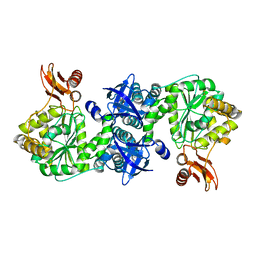 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in the apo-form | | Descriptor: | Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
2DKC
 
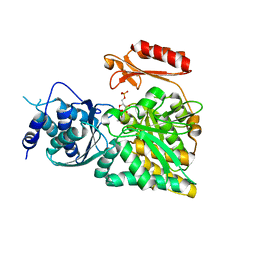 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the substrate complex | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2DKD
 
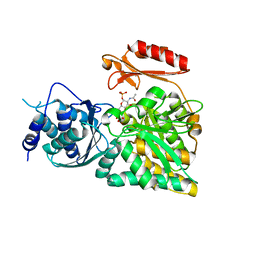 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the product complex | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-galactopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2DKA
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the apo-form | | Descriptor: | Phosphoacetylglucosamine mutase | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
3VLU
 
 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with saturated trimannuronate | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3VLV
 
 | | Crystal structure of Sphingomonas sp. A1 alginate-binding ptotein AlgQ1 in complex with unsaturated triguluronate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlgQ1, CALCIUM ION | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3VLW
 
 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with mannuronate-guluronate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, GLYCEROL, ... | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3A12
 
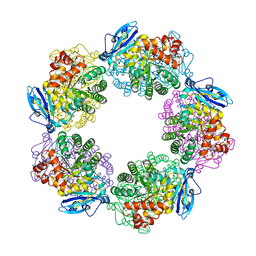 | | Crystal structure of Type III Rubisco complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
3A13
 
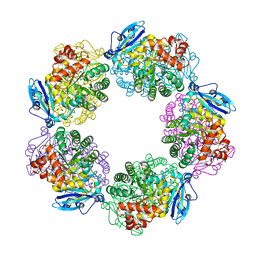 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP and activated with Ca | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based optimization of a Type III Rubisco from a hyperthermophile
To be Published
|
|
5XSW
 
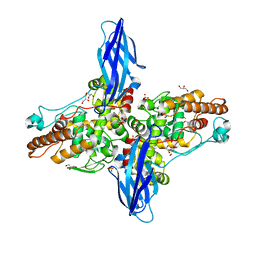 | |
5XSX
 
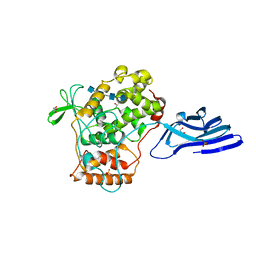 | | Crystal structure of an archaeal chitinase in the substrate-complex form (P212121) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Nishitani, Y, Miki, K. | | Deposit date: | 2017-06-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of an archaeal chitinase ChiD and its ligand complexes.
Glycobiology, 28, 2018
|
|
5XSV
 
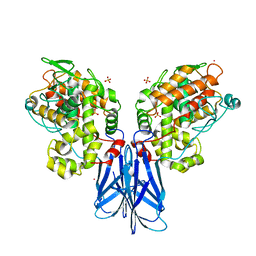 | |
4TQV
 
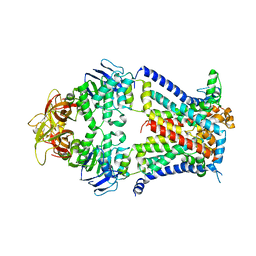 | | Crystal structure of a bacterial ABC transporter involved in the import of the acidic polysaccharide alginate | | Descriptor: | AlgM1, AlgM2, AlgS | | Authors: | Maruyama, Y, Itoh, T, Kaneko, A, Nishitani, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2014-06-12 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.504 Å) | | Cite: | Structure of a Bacterial ABC Transporter Involved in the Import of an Acidic Polysaccharide Alginate
Structure, 23, 2015
|
|
4TQU
 
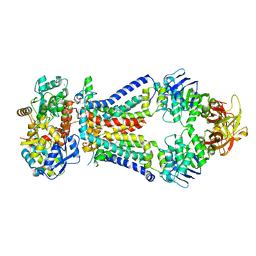 | | Crystal structure of a bacterial ABC transporter involved in the import of the acidic polysaccharide alginate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgM1, AlgM2, ... | | Authors: | Maruyama, Y, Itoh, T, Kaneko, A, Nishitani, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2014-06-12 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Structure of a Bacterial ABC Transporter Involved in the Import of an Acidic Polysaccharide Alginate
Structure, 23, 2015
|
|
5HWS
 
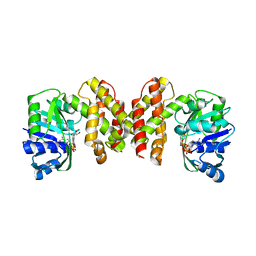 | |
5IJA
 
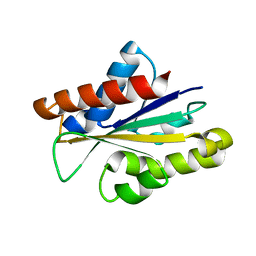 | |
7DVU
 
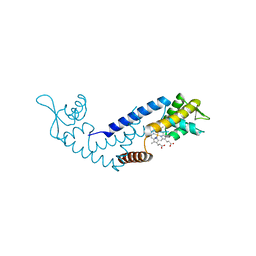 | | Crystal structure of heme sensor protein PefR in complex with heme and cyanide | | Descriptor: | CYANIDE ION, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVT
 
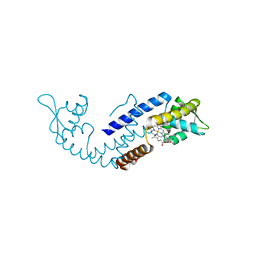 | | Crystal structure of heme sensor protein PefR in complex with heme and carbon monoxide | | Descriptor: | CARBON MONOXIDE, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVR
 
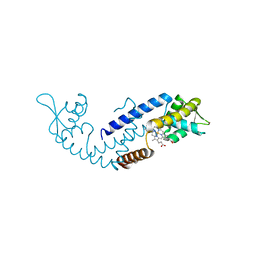 | | Crystal structure of heme sensor protein PefR from Streptococcus agalactiae in complex with heme | | Descriptor: | COBALT (II) ION, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVV
 
 | | Heme sensor protein PefR from Streptococcus agalactiae bound to operator DNA (28-mer) | | Descriptor: | DNA (28-MER), HTH marR-type domain-containing protein | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
