5G26
 
 | | Unveiling the Mechanism Behind the in-meso Crystallization of Membrane Proteins | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, INTIMIN | | Authors: | Zabara, A, Newman, J, Peat, T.S. | | Deposit date: | 2016-04-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Nanoscience Behind the Art of in-Meso Crystallization of Membrane Proteins.
Nanoscale, 9, 2017
|
|
4ZF7
 
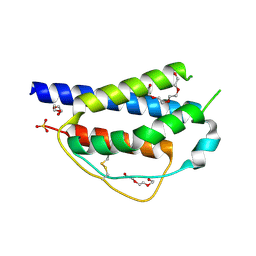 | | Crystal structure of ferret interleukin-2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Interleukin 2, PENTAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Newman, J, McKinstry, W.J, Adams, T.E. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural and functional characterisation of ferret interleukin-2.
Dev.Comp.Immunol., 55, 2015
|
|
5HY2
 
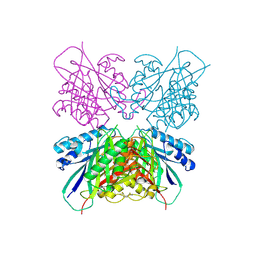 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HXZ
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HY1
 
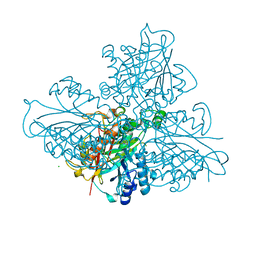 | | high resolution structure of barbiturase | | Descriptor: | 1,3,5-triazine-2,4,6-triol, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Scott, C, Balotra, S, Wilding, M, Newman, J. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HXU
 
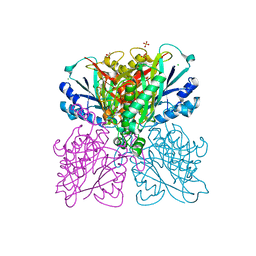 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5AKQ
 
 | | X-ray structure and mutagenesis studies of the N-isopropylammelide isopropylaminohydrolase, AtzC | | Descriptor: | CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ZINC ION | | Authors: | Balotra, S, Warden, A.C, Newman, J, Briggs, L.J, Scott, C, Peat, T.S. | | Deposit date: | 2015-03-05 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
5BK6
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-12 | | Release date: | 2018-02-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
5HY0
 
 | | orotic acid hydrolase | | Descriptor: | GLYCEROL, Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5A25
 
 | | Rational engineering of a mesophilic carbonic anhydrase to an extreme halotolerant biocatalyst | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, SODIUM ION, ... | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2015-05-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
5HY4
 
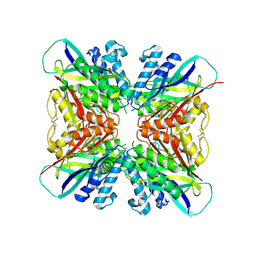 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
6R7T
 
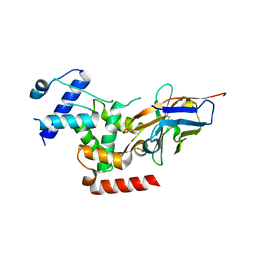 | | Crystal Structure of human Melanoma-associated antigen B1 (MAGEB1) in complex with nanobody | | Descriptor: | Melanoma-associated antigen B1, anti MAGEB1 nanobody | | Authors: | Ye, M, Newman, J, Pike, A.C.W, Burgess-Brown, N, Cooper, C.D.O, Bountra, C, Arrowsmith, C, Edwards, A, Gileadi, O, von Delft, F. | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Crystal Structure of Melanoma-associated antigen B1 (MAGEB1) in complex with nanobody
To Be Published
|
|
6PII
 
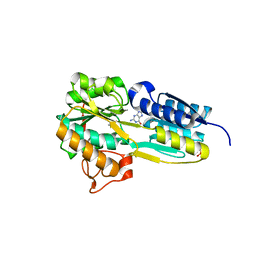 | | The evolving story of AtzT, a periplasmic binding protein | | Descriptor: | Atrazine periplasmic binding protein, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | Deposit date: | 2019-06-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PI5
 
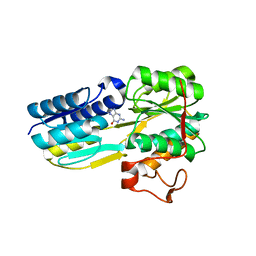 | | The evolving story of AtzT, a periplasmic binding protein | | Descriptor: | Atrazine periplasmic binding protein, DIMETHYL SULFOXIDE, GUANINE | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | Deposit date: | 2019-06-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PI6
 
 | | The evolving story of AtzT, a periplasmic binding protein | | Descriptor: | 4-(ethylamino)-6-[(propan-2-yl)amino]-1,3,5-triazin-2-ol, Atrazine periplasmic binding protein, DIMETHYL SULFOXIDE | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | Deposit date: | 2019-06-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8DFY
 
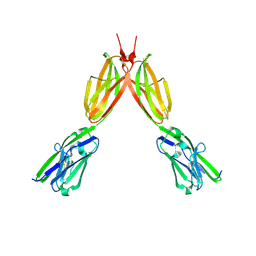 | | Crystal structure of Human BTN2A1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, ... | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.552 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
8DFX
 
 | | Crystal structure of Human BTN2A1-BTN3A1 Ectodomain Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1 | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.55 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
8DFW
 
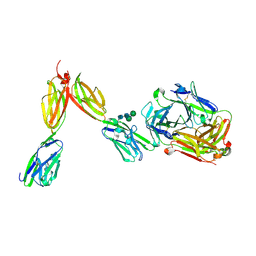 | | Crystal Structure of Human BTN2A1 in Complex With Vgamma9-Vdelta2 T Cell Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, ... | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
6D63
 
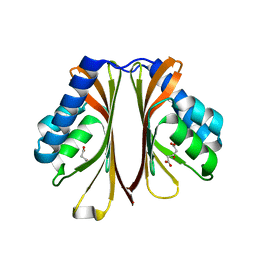 | | The structure of AtzH: a little known member of the atrazine breakdown pathway | | Descriptor: | 3-oxopentanedioic acid, atzH | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L. | | Deposit date: | 2018-04-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel decarboxylating amidohydrolase involved in avoiding metabolic dead ends during cyanuric acid catabolism in Pseudomonas sp. strain ADP.
PLoS ONE, 13, 2018
|
|
3ZSX
 
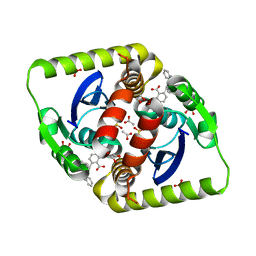 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-({[2-(benzylcarbamoyl)benzyl](prop-2-en-1-yl)amino}methyl)-1,3-benzodioxole-4-carboxylate, ACETATE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZSV
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (R)-[(2S)-5-carboxy-2-(2-carboxyethyl)-2,3-dihydro-1,4-benzodioxin-6-yl]methyl-[[2-[(4-methoxyphenyl)methylcarbamoyl]phenyl]methyl]-prop-2-enyl-azanium, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZSY
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (R)-(4-CARBOXY-1,3-BENZODIOXOL-5-YL)METHYL-[[2-(CYCLOHEXYLMETHYLCARBAMOYL)PHENYL]METHYL]-METHYL-AZANIUM, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZSZ
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (R)-[2-[[(2S)-BUTAN-2-YL]CARBAMOYL]PHENYL]METHYL-[(4-CARBOXY-1,3-BENZODIOXOL-5-YL)METHYL]-METHYL-AZANIUM, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZT1
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (4-CARBOXY-1,3-BENZODIOXOL-5-YL)METHYL-[[2-[(4-METHOXYPHENYL)METHYLCARBAMOYL]PHENYL]METHYL]AZANIUM, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZT2
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 5-[(E)-(2-OXO-2,3-DIHYDRO-1H-INDEN-1-YLIDENE)METHYL]-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
