8HH8
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH6
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,step waiting,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH7
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3, 81 degrees, lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH5
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,120 degrees,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH2
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HHC
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd',lowATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HHB
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,step waiting,lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HHA
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,120 degrees,lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH9
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3, 90 degrees, low ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH4
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,101 degrees, highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH1
 
 | | FoF1-ATPase from Bacillus PS3, 81 degrees, highATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH3
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,90 degrees,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
7XKH
 
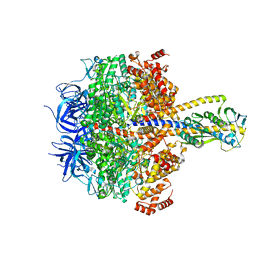 | | Nucleotide-depleted F1 domain of FoF1-ATPase from Bacillus PS3, state1 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKO
 
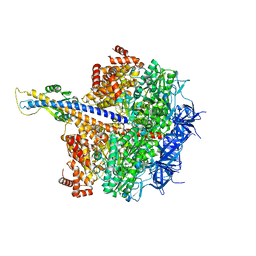 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,nucleotide depeleted | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKQ
 
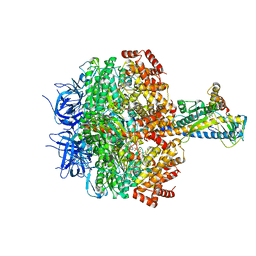 | | F1 domain of FoF1-ATPase with the down form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKR
 
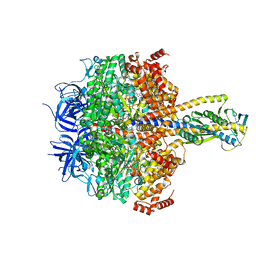 | | F1 domain of FoF1-ATPase with the up form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKP
 
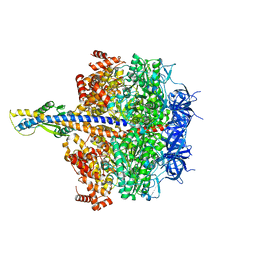 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,unisite condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
2A6W
 
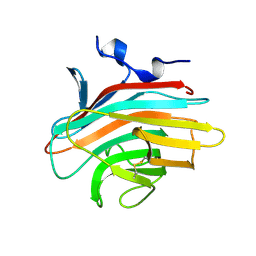 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), metal-free form | | Descriptor: | Emp46p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A71
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), orthorhombic crystal form | | Descriptor: | Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Y
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), tetragonal crystal form | | Descriptor: | Emp47p (form1), SULFATE ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6V
 
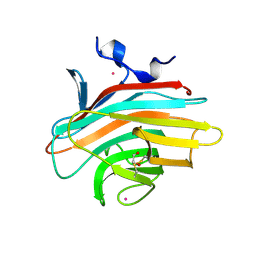 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), potassium-bound form | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Z
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 1 | | Descriptor: | Emp47p (form2) | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6X
 
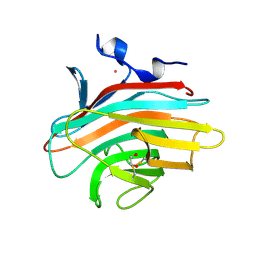 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), Y131F mutant | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A70
 
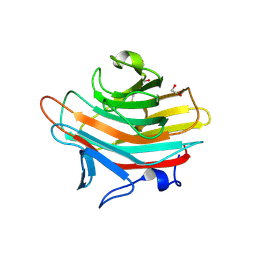 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Katoh, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
7VAX
 
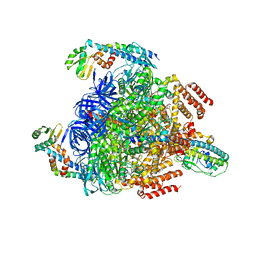 | | V1EG of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state1-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2021-08-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
