2FYG
 
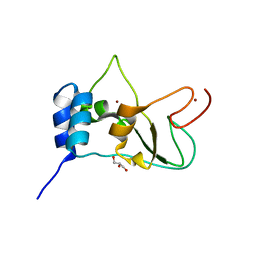 | | Crystal structure of NSP10 from Sars coronavirus | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Joseph, J.S, Saikatendu, K.S, Subramanian, V, Neuman, B.W, Brooun, A, Griffith, M, Moy, K, Yadav, M.K, Velasquez, J, Buchmeier, M.J, Stevens, R.C, Kuhn, P. | | Deposit date: | 2006-02-07 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs.
J.Virol., 80, 2006
|
|
3U2J
 
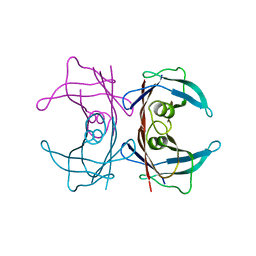 | | Neutron crystal structure of human Transthyretin | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3U2I
 
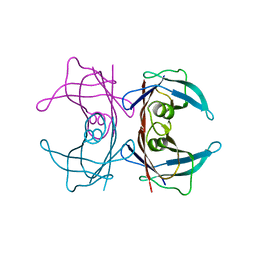 | | X-ray crystal structure of human Transthyretin at room temperature | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3VXF
 
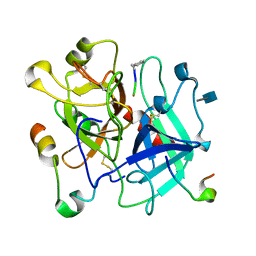 | | X/N Joint refinement of Human alpha-thrombin-Bivalirudin complex PD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.602 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
3VXE
 
 | | Human alpha-thrombin-Bivalirudin complex at PD5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
1VJ2
 
 | |
1VKY
 
 | |
1VKB
 
 | |
1VLR
 
 | |
1VKM
 
 | |
1VJ1
 
 | |
1J5S
 
 | |
1J6U
 
 | |
1O58
 
 | |
1O5U
 
 | |
1O4U
 
 | |
1O50
 
 | |
1O1X
 
 | |
1O1Y
 
 | |
1O20
 
 | |
1O4S
 
 | |
1O4T
 
 | |
1O0X
 
 | |
1O51
 
 | |
1O59
 
 | |
