1URS
 
 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-11-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins
J.Mol.Biol., 335, 2004
|
|
1USG
 
 | | L-leucine-binding protein, apo form | | Descriptor: | LEUCINE-SPECIFIC BINDING PROTEIN | | Authors: | Magnusson, U, Salopek-Sondi, B, Luck, L.A, Mowbray, S.L. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structures of the Leucine-Binding Protein Illustrate Conformational Changes and the Basis of Ligand Specificity
J.Biol.Chem., 279, 2004
|
|
1URD
 
 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius provide insight into acid stability of proteins | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1USL
 
 | | Structure Of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, Complexed With Phosphate. | | Descriptor: | PHOSPHATE ION, RIBOSE 5-PHOSPHATE ISOMERASE B | | Authors: | Roos, A.K, Andersson, C.E, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-11-25 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase Has a Known Fold, But a Novel Active Site
J.Mol.Biol., 335, 2004
|
|
1URG
 
 | | X-ray structures from the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1USK
 
 | | L-leucine-binding protein with leucine bound | | Descriptor: | LEUCINE, LEUCINE-SPECIFIC BINDING PROTEIN | | Authors: | Magnusson, U, Salopek-Sondi, B, Luck, L.A, Mowbray, S.L. | | Deposit date: | 2003-11-25 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structures of the Leucine-Binding Protein Illustrate Conformational Changes and the Basis of Ligand Specificity
J.Biol.Chem., 279, 2004
|
|
1USI
 
 | | L-leucine-binding protein with phenylalanine bound | | Descriptor: | LEUCINE-SPECIFIC BINDING PROTEIN, PHENYLALANINE | | Authors: | Magnusson, U, Salopek-Sondi, B, Luck, L.A, Mowbray, S.L. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Leucine-Binding Protein Illustrate Conformational Changes and the Basis of Ligand Specificity
J.Biol.Chem., 279, 2004
|
|
1W74
 
 | | X-ray structure of peptidyl-prolyl cis-trans isomerase A, PpiA, Rv0009, from Mycobacterium tuberculosis. | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Henriksson, L.M, Johansson, P, Unge, T, Mowbray, S.L. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of Peptidyl-Prolyl Cis-Trans Isomerase a from Mycobacterium Tuberculosis
Eur.J.Biochem., 271, 2004
|
|
1YLK
 
 | | Crystal Structure of Rv1284 from Mycobacterium tuberculosis in Complex with Thiocyanate | | Descriptor: | Hypothetical protein Rv1284/MT1322, THIOCYANATE ION, ZINC ION | | Authors: | Covarrubias, A.S, Larsson, A.M, Hogbom, M, Lindberg, J, Bergfors, T, Bjorkelid, C, Mowbray, S.L, Unge, T, Jones, T.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-19 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of carbonic anhydrases from Mycobacterium tuberculosis.
J.Biol.Chem., 280, 2005
|
|
1YM3
 
 | | Crystal Structure of carbonic anhydrase RV3588c from Mycobacterium tuberculosis | | Descriptor: | CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), MAGNESIUM ION, ZINC ION | | Authors: | Covarrubias, A.S, Larsson, A.M, Hogbom, M, Lindberg, J, Bergfors, T, Bjorkelid, C, Mowbray, S.L, Unge, T, Jones, T.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and function of carbonic anhydrases from Mycobacterium tuberculosis.
J.Biol.Chem., 280, 2005
|
|
2BVC
 
 | | Crystal structure of Mycobacterium tuberculosis glutamine synthetase in complex with a transition state mimic | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLUTAMINE SYNTHETASE 1, ... | | Authors: | Krajewski, W.W, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2005-06-23 | | Release date: | 2005-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Mycobacterium Tuberculosis Glutamine Synthetase in Complex with a Transition-State Mimic Provides Functional Insights.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BNG
 
 | | Structure of an M.tuberculosis LEH-like epoxide hydrolase | | Descriptor: | CALCIUM ION, MB2760 | | Authors: | Johansson, P, Arand, M, Unge, T, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2005-03-24 | | Release date: | 2005-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an Atypical Epoxide Hydrolase from Mycobacterium Tuberculosis Gives Insights Into its Function.
J.Mol.Biol., 351, 2005
|
|
2D1F
 
 | | Structure of Mycobacterium tuberculosis threonine synthase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine synthase | | Authors: | Covarrubias, A.S, Bergfors, T, Mannerstedt, K, Oscarson, S, Jones, T.A, Mowbray, S.L, Hogbom, M. | | Deposit date: | 2005-08-20 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, biochemical, and in vivo investigations of the threonine synthase from Mycobacterium tuberculosis.
J.Mol.Biol., 381, 2008
|
|
2C82
 
 | | X-Ray Structure Of 1-Deoxy-D-xylulose 5-phosphate Reductoisomerase, DXR, Rv2870c, From Mycobacterium tuberculosis | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, SULFATE ION | | Authors: | Henriksson, L.M, Bjorkelid, C, Mowbray, S.L, Unge, T. | | Deposit date: | 2005-11-30 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A Resolution Structure of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase, a Potential Drug Target.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3GA5
 
 | | X-ray structure of glucose/galactose receptor from Salmonella typhimurium in complex with (2R)-glyceryl-beta-D-galactopyranoside | | Descriptor: | (2R)-2,3-dihydroxypropyl beta-D-galactopyranoside, CALCIUM ION, D-galactose-binding periplasmic protein, ... | | Authors: | Sooriyaarachchi, S, Ubhayasekera, W, Mowbray, S.L. | | Deposit date: | 2009-02-16 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structure of glucose/galactose receptor from Salmonella typhimurium in complex with the physiological ligand, (2R)-glyceryl-beta-D-galactopyranoside
Febs J., 276, 2009
|
|
3HBE
 
 | |
3HBD
 
 | |
3HBH
 
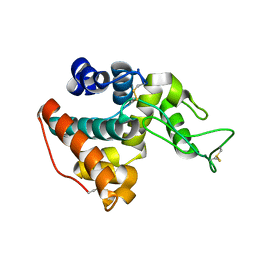 | |
3K8C
 
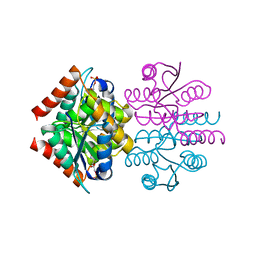 | |
3K7S
 
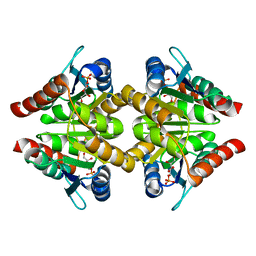 | |
3K7O
 
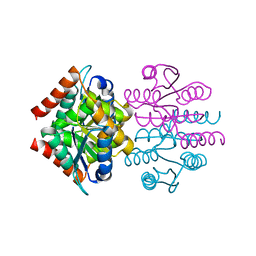 | |
3K7P
 
 | |
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
1NWW
 
 | | Limonene-1,2-epoxide hydrolase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEPTANAMIDE, Limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
1NU3
 
 | | Limonene-1,2-epoxide hydrolase in complex with valpromide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PROPYLPENTANAMIDE, limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
