2D1I
 
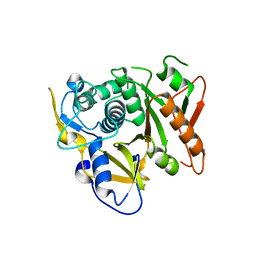 | | Structure of human Atg4b | | Descriptor: | Cysteine protease APG4B | | Authors: | Kumanomidou, T, Mizushima, T, Komatsu, M, Suzuki, A, Tanida, I, Sou, Y.S, Ueno, T, Kominami, E, Tanaka, K, Yamane, T. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Atg4b, a Processing and De-conjugating Enzyme for Autophagosome-forming Modifiers
J.Mol.Biol., 355, 2006
|
|
2DYS
 
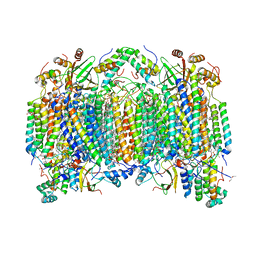 | | Bovine heart cytochrome C oxidase modified by DCCD | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
2DYR
 
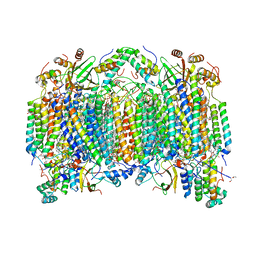 | | Bovine heart cytochrome C oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
3WDZ
 
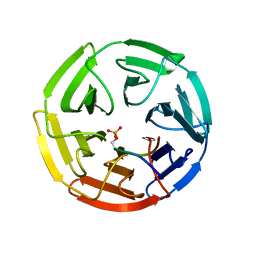 | | Crystal Structure of Keap1 in Complex with phosphorylated p62 | | Descriptor: | Kelch-like ECH-associated protein 1, Peptide from Sequestosome-1 | | Authors: | Fukutomi, T, Takagi, K, Mizushima, T, Tanaka, K, Komatsu, M, Yamamoto, M. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy.
Mol.Cell, 51, 2013
|
|
3A4U
 
 | | Crystal structure of MCFD2 in complex with carbohydrate recognition domain of ERGIC-53 | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Nishio, M, Kamiya, Y, Mizushima, T, Wakatsuki, S, Sasakawa, H, Yamamoto, K, Uchiyama, S, Noda, M, McKay, A.R, Fukui, K, Hauri, H.P, Kato, K. | | Deposit date: | 2009-07-17 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3WSO
 
 | | Crystal structure of the Skp1-FBG3 complex | | Descriptor: | F-box only protein 44, S-phase kinase-associated protein 1 | | Authors: | Kumanomidou, T, Nishio, K, Takagi, K, Nakagawa, T, Suzuki, A, Yamane, T, Tokunaga, F, Iwai, K, Murakami, A, Yoshida, Y, Tanaka, K, Mizushima, T. | | Deposit date: | 2014-03-18 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Differences between a Glycoprotein Specific F-Box Protein Fbs1 and Its Homologous Protein FBG3
Plos One, 10, 2015
|
|
2B9U
 
 | | Crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase from sulfolobus tokodaii | | Descriptor: | hypothetical dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Rajakannan, V, Kondo, K, Mizushima, T, Suzuki, A, Yamane, T. | | Deposit date: | 2005-10-13 | | Release date: | 2006-10-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase from sulfolobus tokodaii
TO BE PUBLISHED
|
|
2EFX
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine amide | | Descriptor: | BARIUM ION, D-amino acid amidase, PHENYLALANINE AMIDE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EFU
 
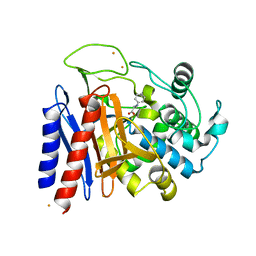 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine | | Descriptor: | BARIUM ION, D-Amino acid amidase, PHENYLALANINE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3VR0
 
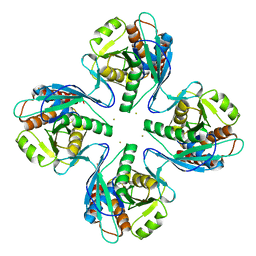 | | Crystal structure of Pyrococcus furiosus PbaB, an archaeal proteasome activator | | Descriptor: | GOLD ION, Putative uncharacterized protein | | Authors: | Kumoi, K, Satoh, T, Hiromoto, T, Mizushima, T, Kamiya, Y, Noda, M, Uchiyama, S, Murata, K, Yagi, H, Kato, K. | | Deposit date: | 2012-04-02 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An archaeal homolog of proteasome assembly factor functions as a proteasome activator
Plos One, 8, 2013
|
|
3WHJ
 
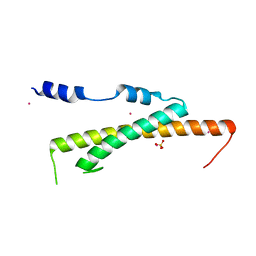 | | Crystal structure of Nas2 N-terminal domain | | Descriptor: | CADMIUM ION, Probable 26S proteasome regulatory subunit p27, SULFATE ION | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHK
 
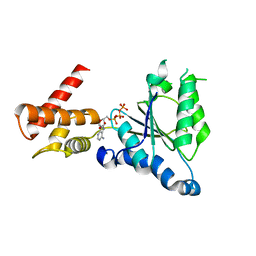 | | Crystal structure of PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHL
 
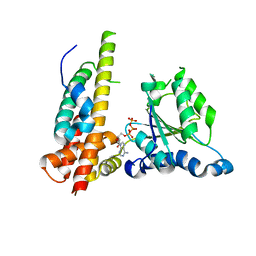 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WN7
 
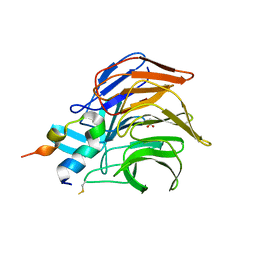 | | Crystal Structure of Keap1 in Complex with the N-terminal region of the Nrf2 transcription factor | | Descriptor: | ACETATE ION, Kelch-like ECH-associated protein 1, Peptide from Nuclear factor erythroid 2-related factor 2 | | Authors: | Fukutomi, T, Takagi, K, Mizushima, T, Ohuchi, N, Yamamoto, M. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1
Mol.Cell.Biol., 34, 2014
|
|
3X3H
 
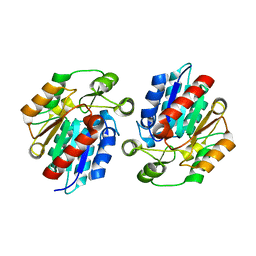 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) 3KP (K176P, K199P, K224P) triple mutant | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H, Okazaki, S. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
TO BE PUBLISHED
|
|
3W31
 
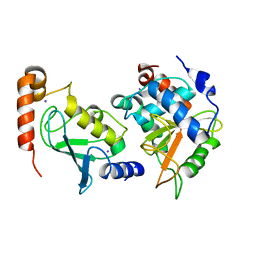 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | IODIDE ION, ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
J.Mol.Biol., 425, 2013
|
|
3W30
 
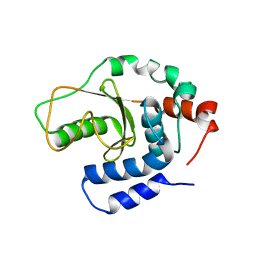 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | ORF169b | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Basis for the Recognition of Ubc13 by the Shigella flexneri Effector OspI.
J.Mol.Biol., 425, 2013
|
|
3VLE
 
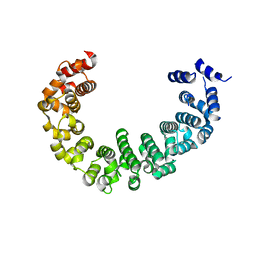 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLD
 
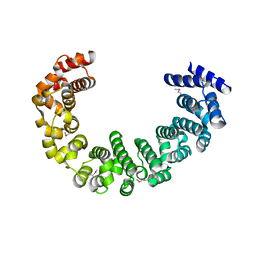 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLF
 
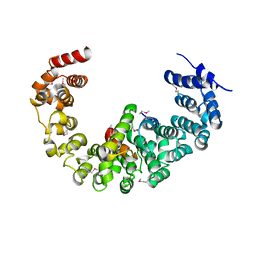 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VL1
 
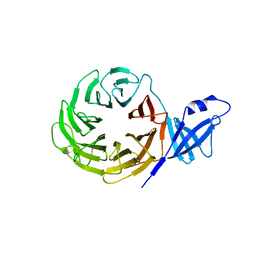 | | Crystal structure of yeast Rpn14 | | Descriptor: | 26S proteasome regulatory subunit RPN14 | | Authors: | Kim, S, Nishide, A, Saeki, Y, Takagi, K, Tanaka, K, Kato, K, Mizushima, T. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3WRI
 
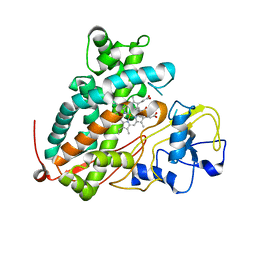 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of P450cam intermediate
To be Published
|
|
3WRM
 
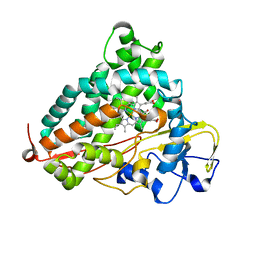 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of P450cam intermedite
To be published
|
|
3WRJ
 
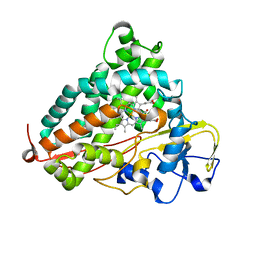 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of P450cam intermedite
to be published
|
|
3WXR
 
 | | Yeast 20S proteasome with a mutation of alpha7 subunit | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Yashiroda, H, Toda, Y, Otsu, S, Takagi, K, Mizushima, T, Murata, S. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | N-terminal alpha 7 deletion of the proteasome 20S core particle substitutes for yeast PI31 function
Mol.Cell.Biol., 35, 2015
|
|
