3AJC
 
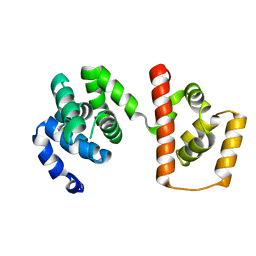 | | Structure of the MC domain of FliG (PEV), a CW-biased mutant | | Descriptor: | Flagellar motor switch protein fliG | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the rotational switching mechanism of the bacterial flagellar motor
Plos Biol., 9, 2011
|
|
5H72
 
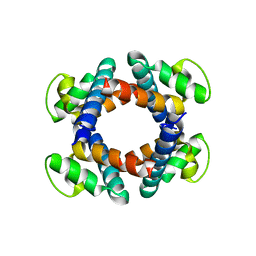 | | Structure of the periplasmic domain of FliP | | Descriptor: | Flagellar biosynthetic protein FliP | | Authors: | Fukumura, T, Kawaguchi, T, Saijo-Hamano, Y, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-11-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
PLoS Biol., 15, 2017
|
|
6JY0
 
 | | CryoEM structure of S.typhimurium R-type straight flagellar filament made of FljB (A461V) | | Descriptor: | Flagellin | | Authors: | Yamaguchi, T, Toma, S, Terahara, N, Miyata, T, Minamino, T, Ashikara, M, Namba, K, Kato, T. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural and Functional Comparison ofSalmonellaFlagellar Filaments Composed of FljB and FliC.
Biomolecules, 10, 2020
|
|
9IWQ
 
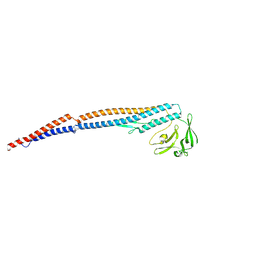 | | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament | | Descriptor: | Flagellin | | Authors: | Waraich, K, Makino, F, Miyata, T, Kinoshita, M, Minamino, T, Namba, K. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament
To Be Published
|
|
5B0O
 
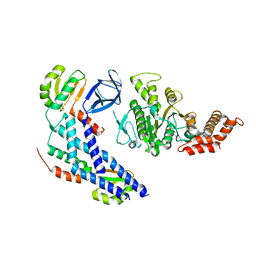 | | Structure of the FliH-FliI complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Flagellar assembly protein FliH, Flagellum-specific ATP synthase | | Authors: | Imada, K, Uchida, Y, Kinoshita, M, Namba, K, Minamino, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2DPY
 
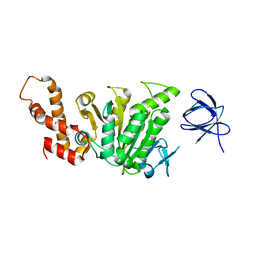 | |
3AJW
 
 | | Structure of FliJ, a soluble component of flagellar type III export apparatus | | Descriptor: | Flagellar fliJ protein, MERCURY (II) ION | | Authors: | Imada, K, Ibuki, T, Minamino, T, Namba, K. | | Deposit date: | 2010-06-23 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases
Nat.Struct.Mol.Biol., 18, 2011
|
|
5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
6AI2
 
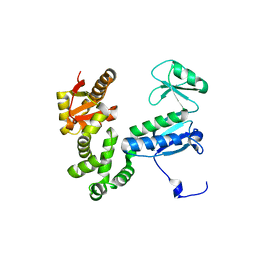 | |
6AI1
 
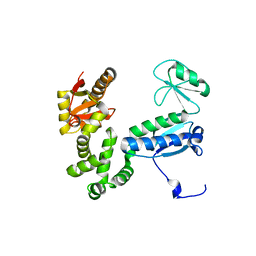 | |
6AI3
 
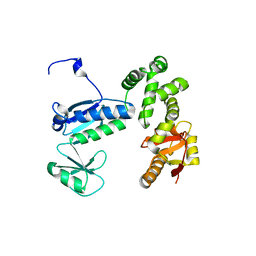 | |
6AI0
 
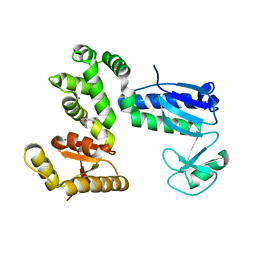 | | Structure of the 328-692 fragment of FlhA (orthorhombic form) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, Flagellar biosynthesis protein FlhA | | Authors: | Ogawa, Y, Kinoshita, M, Minamino, T, Imada, K. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Substrate Specificity Switch Mechanism of the Type III Protein Export Apparatus.
Structure, 27, 2019
|
|
7CLR
 
 | | CryoEM structure of S.typhimurium flagellar LP ring | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein | | Authors: | Yamaguchi, T, Makino, F, Miyata, T, Minamino, T, Kato, T, Namba, K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the molecular bushing of the bacterial flagellar motor.
Nat Commun, 12, 2021
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7CTN
 
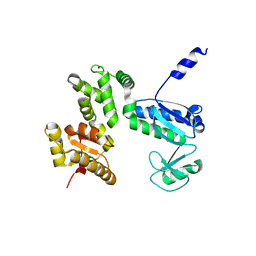 | | Structure of the 328-692 fragment of FlhA (E351A/D356A) | | Descriptor: | Flagellar biosynthesis protein FlhA | | Authors: | Kida, M, Takekawa, N, Kinoshita, M, Inoue, Y, Minamino, T, Imada, K. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The FlhA linker mediates flagellar protein export switching during flagellar assembly.
Commun Biol, 4, 2021
|
|
3A7M
 
 | | Structure of FliT, the flagellar type III chaperone for FliD | | Descriptor: | Flagellar protein fliT | | Authors: | Imada, K, Minamino, T, Kinoshita, M, Namba, K. | | Deposit date: | 2009-09-29 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A5I
 
 | |
8Z4D
 
 | | Structure of the S-ring region of the Vibrio flagellar MS-ring protein FliF with 34-fold symmetry applied | | Descriptor: | Flagellar M-ring protein,Flagellar motor switch protein FliG | | Authors: | Takekawa, N, Nishikino, T, Kishikawa, J, Hirose, M, Kato, T, Imada, K, Homma, M. | | Deposit date: | 2024-04-17 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural analysis of S-ring composed of FliFG fusion proteins in marine Vibrio polar flagellar motors
To Be Published
|
|
8Z4G
 
 | | Structure of the S-ring region of the Vibrio flagellar MS-ring protein FliF with 35-fold symmetry applied | | Descriptor: | Flagellar M-ring protein,Flagellar motor switch protein FliG | | Authors: | Takekawa, N, Nishikino, T, Kishikawa, J, Hirose, M, Kato, T, Imada, K, Homma, M. | | Deposit date: | 2024-04-17 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural analysis of S-ring composed of FliFG fusion proteins in marine Vibrio polar flagellar motors
To Be Published
|
|
5WRH
 
 | |
5XAM
 
 | |
5XAP
 
 | | Crystal structure of SecDF in I form (C2 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Protein translocase subunit SecD | | Authors: | Tsukazaki, T, Tanaka, Y, Furukwa, A. | | Deposit date: | 2017-03-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Tunnel Formation Inferred from the I-Form Structures of the Proton-Driven Protein Secretion Motor SecDF
Cell Rep, 19, 2017
|
|
5XAN
 
 | | Crystal structure of SecDF in I form (P212121 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POLYETHYLENE GLYCOL (N=34), Protein translocase subunit SecD | | Authors: | Tsukazaki, T, Tanaka, Y, Furukwa, A. | | Deposit date: | 2017-03-14 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Tunnel Formation Inferred from the I-Form Structures of the Proton-Driven Protein Secretion Motor SecDF
Cell Rep, 19, 2017
|
|
7CIK
 
 | | Structure of the 58-213 fragment of FliF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Flagellar M-ring protein | | Authors: | Takekawa, T, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Two Distinct Conformations in 34 FliF Subunits Generate Three Different Symmetries within the Flagellar MS-Ring.
Mbio, 12, 2021
|
|
2ZVY
 
 | |
