8E66
 
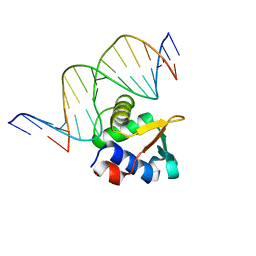 | | ETV6 H396Y variant bound to DNA containing the sequence GGAA | | 分子名称: | CACODYLATE ION, Complementary 15 bp strand, GGAA-containing 15 bp DNA, ... | | 著者 | Scheu, K, Chan, A.C, Murphy, M.E, McIntosh, L.P. | | 登録日 | 2022-08-22 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The functional role of histidine within the ETV6 ETS domain
to be published
|
|
8E67
 
 | |
7JU2
 
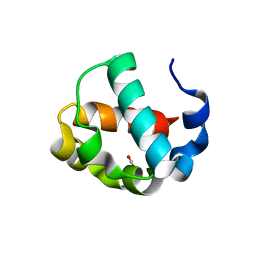 | | Crystal structure of the monomeric ETV6 PNT domain | | 分子名称: | FORMIC ACID, Transcription factor ETV6 | | 著者 | Gerak, C.A.N, Kolesnikov, M, Murphy, M.E.P, McIntosh, L.P. | | 登録日 | 2020-08-19 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85002184 Å) | | 主引用文献 | Biophysical characterization of the ETV6 PNT domain polymerization interfaces.
J.Biol.Chem., 296, 2021
|
|
1ULP
 
 | |
1ULO
 
 | |
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | 登録日 | 2010-01-08 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
1C5H
 
 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | 分子名称: | ENDO-1,4-BETA-XYLANASE | | 著者 | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | 登録日 | 1999-11-24 | | 公開日 | 2000-05-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
1C5I
 
 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | 分子名称: | ENDO-1,4-BETA-XYLANASE, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | 著者 | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | 登録日 | 1999-11-24 | | 公開日 | 2000-05-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
6CAH
 
 | |
1CX1
 
 | | SECOND N-TERMINAL CELLULOSE-BINDING DOMAIN FROM CELLULOMONAS FIMI BETA-1,4-GLUCANASE C, NMR, 22 STRUCTURES | | 分子名称: | ENDOGLUCANASE C | | 著者 | Brun, E, Johnson, P.E, Creagh, L.A, Haynes, C.A, Tomme, P, Webster, P, Kilburn, D.G, McIntosh, L.P. | | 登録日 | 1999-08-27 | | 公開日 | 2000-04-02 | | 最終更新日 | 2017-02-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and binding specificity of the second N-terminal cellulose-binding domain from Cellulomonas fimi endoglucanase C.
Biochemistry, 39, 2000
|
|
2WQQ
 
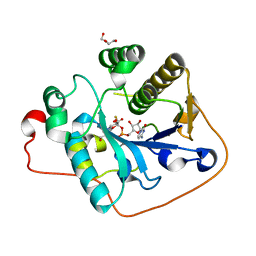 | | Crystallographic analysis of monomeric CstII | | 分子名称: | ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER | | 著者 | Chan, P.H.W, Lairson, L.L, Lee, H.J, Wakarchuk, W.W, Strynadka, N.C.J, Withers, S.G, McIntosh, L.P. | | 登録日 | 2009-08-25 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | NMR Spectroscopic Characterization of the Sialyltransferase Cstii from Camplyobacter Jejuni: Histidine 188 is the General Base.
Biochemistry, 48, 2009
|
|
6DA1
 
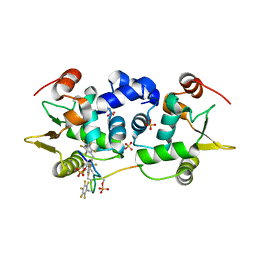 | | ETS1 in complex with synthetic SRR mimic | | 分子名称: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | 著者 | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | 登録日 | 2018-05-01 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.000127 Å) | | 主引用文献 | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6DAT
 
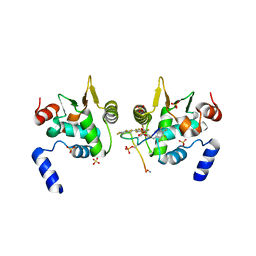 | | ETS1 in complex with synthetic SRR mimic | | 分子名称: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | 著者 | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | 登録日 | 2018-05-02 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.35002637 Å) | | 主引用文献 | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6XFK
 
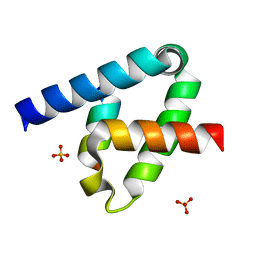 | | Crystal structure of the type III secretion system pilotin-secretin complex InvH-InvG | | 分子名称: | SULFATE ION, Type 3 secretion system pilotin, Type 3 secretion system secretin | | 著者 | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
1SN8
 
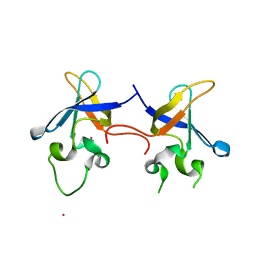 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | 分子名称: | LEAD (II) ION, Ribonuclease E | | 著者 | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | 登録日 | 2004-03-10 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
6XFL
 
 | | Structural characterization of the type III secretion system pilotin-secretin complex InvH-InvG by NMR spectroscopy | | 分子名称: | Type 3 secretion system pilotin, Type 3 secretion system secretin | | 著者 | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
1SMX
 
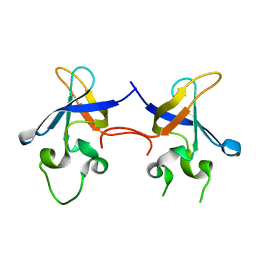 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | 分子名称: | Ribonuclease E | | 著者 | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | 登録日 | 2004-03-09 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
6XFJ
 
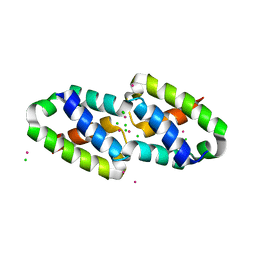 | | Crystal structure of the type III secretion pilotin InvH | | 分子名称: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | 著者 | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
1SXE
 
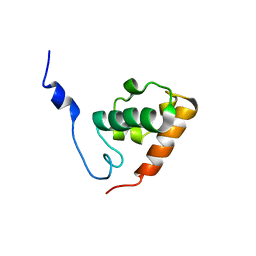 | | The solution structure of the Pointed (PNT) domain from the transcrition factor Erg | | 分子名称: | Transcriptional regulator ERG | | 著者 | Mackereth, C.D, Schaerpf, M, Gentile, L.N, MacIntosh, S.E, Slupsky, C.M, McIntosh, L.P. | | 登録日 | 2004-03-30 | | 公開日 | 2004-09-21 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Diversity in Structure and Function of the Ets Family PNT Domains.
J.Mol.Biol., 342, 2004
|
|
1SXD
 
 | | Solution Structure of the Pointed (PNT) Domain from mGABPa | | 分子名称: | GA repeat binding protein, alpha | | 著者 | Mackereth, C.D, Schaerpf, M, Gentile, L.N, MacIntosh, S.E, Slupsky, C.M, McIntosh, L.P. | | 登録日 | 2004-03-30 | | 公開日 | 2004-09-21 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Diversity in Structure and Function of the Ets Family PNT Domains.
J.Mol.Biol., 342, 2004
|
|
1GGW
 
 | |
3VZL
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | 登録日 | 2012-10-15 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZN
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | 著者 | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | 登録日 | 2012-10-15 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZJ
 
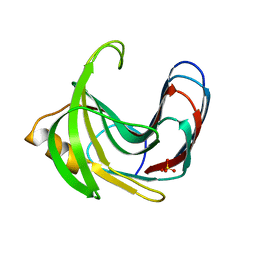 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) E172H mutant | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | 登録日 | 2012-10-14 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.406 Å) | | 主引用文献 | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZK
 
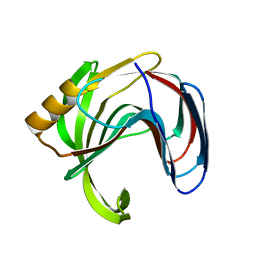 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | 登録日 | 2012-10-14 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
