6P3C
 
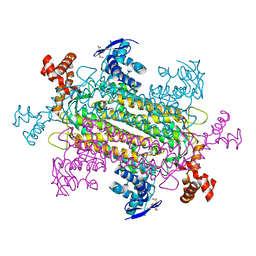 | |
6NZC
 
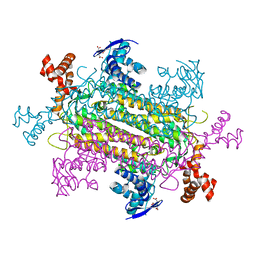 | |
6NZB
 
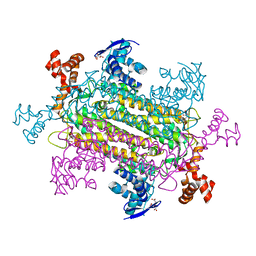 | |
6NZA
 
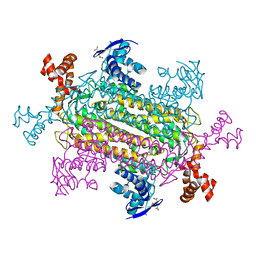 | |
6OS7
 
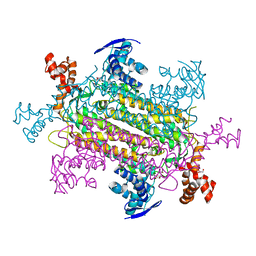 | |
6NZ9
 
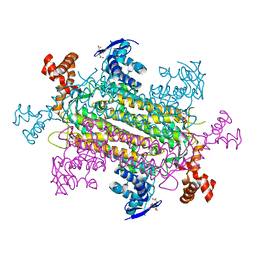 | | Crystal structure of E. coli fumarase C bound to citrate at 1.53 angstrom resolution | | Descriptor: | CITRIC ACID, Fumarate hydratase class II | | Authors: | Stuttgen, G.M, May, J.F, Bhattcharyya, B, Weaver, T.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Closed fumarase C active-site structures reveal SS Loop residue contribution in catalysis.
Febs Lett., 594, 2020
|
|
6E8A
 
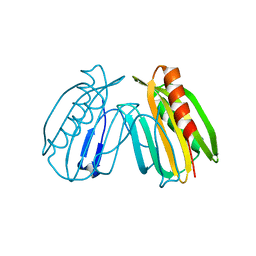 | | Crystal structure of DcrB from Salmonella enterica at 1.92 Angstroms resolution | | Descriptor: | DUF1795 domain-containing protein | | Authors: | Rasmussen, D.M, Soens, R.W, Bhattacharyya, B, May, J.F. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of DcrB, a lipoprotein from Salmonella enterica, reveals flexibility in the N-terminal segment of the Mog1p/PsbP-like fold.
J. Struct. Biol., 204, 2018
|
|
3IVN
 
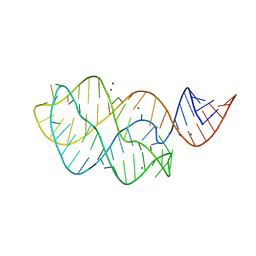 | | Structure of the U65C mutant A-riboswitch aptamer from the Bacillus subtilis pbuE operon | | Descriptor: | A-riboswitch, BROMIDE ION, MAGNESIUM ION | | Authors: | Delfosse, V, Dagenais, P, Chausse, D, Di Tomasso, G, Legault, P. | | Deposit date: | 2009-09-01 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Riboswitch structure: an internal residue mimicking the purine ligand.
Nucleic Acids Res., 38, 2010
|
|
