7P8O
 
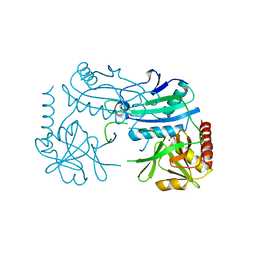 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
8PNW
 
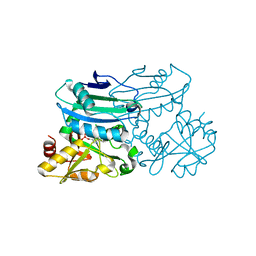 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8PNY
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens complexed with phenylhydrazine and in its apo form | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8AYJ
 
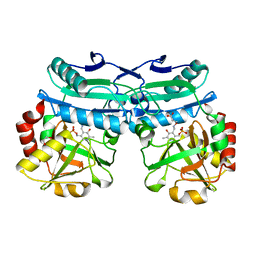 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiens complexed with 3-aminooxypropionic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8AYK
 
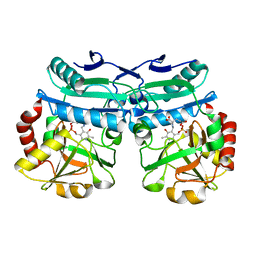 | | Crystal structure of D-amino acid aminotrensferase from Aminobacterium colombiense complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
9UFQ
 
 | | Crystal structure of L-asparaginase from Thermococcus Sibiricus | | Descriptor: | GLYCINE, asparaginase | | Authors: | Matyuta, I.O, Varfolomeeva, L.A, Minyaev, M.E, Dumina, M, Zhdanov, D, Zhgun, A, Pokrovskaya, M, Aleksandrova, S, El'darov, M, Popov, V.O, Boyko, K.M. | | Deposit date: | 2025-04-10 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hyperthermophilic L-Asparaginase from Thermococcus sibiricus and Its Double Mutant with Increased Activity: Insights into Substrate Specificity and Structure.
Int J Mol Sci, 26, 2025
|
|
9UFR
 
 | | Crystal structure of L-asparaginase from Thermococcus Sibiricus double mutation D54G/T56Q | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, GLYCINE, ... | | Authors: | Matyuta, I.O, Varfolomeeva, L.A, Minyaev, M.E, Dumina, M, Zhdanov, D, Zhgun, A, Pokrovskaya, M, Aleksandrova, S, El'darov, M, Popov, V.O, Boyko, K.M. | | Deposit date: | 2025-04-10 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hyperthermophilic L-Asparaginase from Thermococcus sibiricus and Its Double Mutant with Increased Activity: Insights into Substrate Specificity and Structure.
Int J Mol Sci, 26, 2025
|
|
8YRV
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, MAGNESIUM ION | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the Functioning of the D-amino Acid Transaminase from Haliscomenobacter Hydrossis via a Structural and Spectral Analysis of its Complex with 3-Aminooxypropionic Acid.
Acta Naturae, 16, 2024
|
|
8YRT
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis in the holo form obtained at pH 7.0 | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
8YRU
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (apo form) after 15 sec of soaking with phenylhydrazine | | Descriptor: | ACETATE ION, Aminotransferase class IV, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
8AHU
 
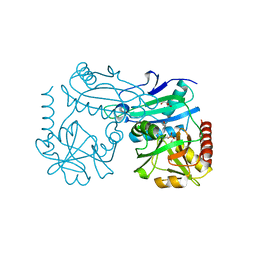 | | Crystal structure of D-amino acid aminotrensferase from Haliscomenobacter hydrossis complexed with D-cycloserine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mechanism of D-Cycloserine Inhibition of D-Amino Acid Transaminase from Haliscomenobacter hydrossis.
Biochemistry Mosc., 88, 2023
|
|
8AIE
 
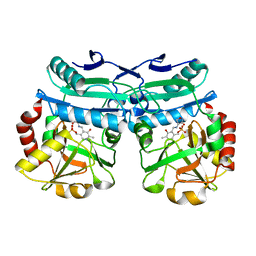 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense complexed with D-cycloserine | | Descriptor: | 3-azanyloxy-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D Structure of D-Аmino Acid Тransaminase from Aminobacterium colombiense in Complex with D-Cycloserine
Crystallography Reports, 68, 2023
|
|
8AHR
 
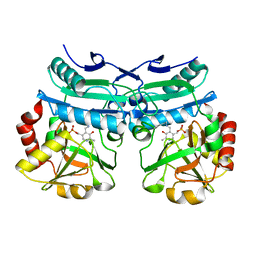 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense in holo form with PLP | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
9J0V
 
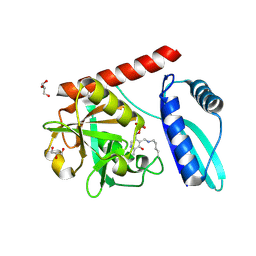 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in P 21 21 21 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dat: predicted D-alanine aminotransferase, GLYCEROL | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | From Structure to Function: Analysis of the First Monomeric Pyridoxal-5'-Phosphate-Dependent Transaminase from the Bacterium Desulfobacula toluolica .
Biomolecules, 14, 2024
|
|
9J0U
 
 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in F 41 3 2 space group | | Descriptor: | Dat: predicted D-alanine aminotransferase | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | From Structure to Function: Analysis of the First Monomeric Pyridoxal-5'-Phosphate-Dependent Transaminase from the Bacterium Desulfobacula toluolica .
Biomolecules, 14, 2024
|
|
9JXU
 
 | | Crystal structure of cysteine synthase A from Limosilactobacillus reuteri LR1 in its apo form | | Descriptor: | 1,2-ETHANEDIOL, Cysteine synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Matyuta, I.O, Les, E.K, Minyaev, M.E, Pometun, A.A, Tishkov, V.I, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-10-11 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-functional examination of novel cysteine synthase A (CysK) from Limosilactobacillus reuteri LR1
To Be Published
|
|
9JLO
 
 | | Crystal structure L-lactate dehydrogenase from Lactobacillus reuteri in its apoform | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase | | Authors: | Matyuta, I.O, Minyaev, M.E, Shirokova, A.A, Pometun, A.A, Tishkov, V.I, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-09-19 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Novel extremely stable and active lactate dehydrogenase from l reuteri: structure-function studies and promising usage as an agent against pathogenic biofilms
To Be Published
|
|
8ONN
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONL
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | Descriptor: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONM
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
To Be Published
|
|
8OQ0
 
 | | Crystal structure of tailspike depolymerase (APK09_gp48) from Acinetobacter phage APK09 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tailspike protein | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Popova, A.V, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii .
Int J Mol Sci, 24, 2023
|
|
8OPZ
 
 | | Crystal structure of a tailspike depolymerase (APK16_gp47) from Acinetobacter phage APK16 | | Descriptor: | GLYCEROL, Tailspike depolymerase (APK16_gp47) from Acinetobacter phage APK16 | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii .
Int J Mol Sci, 24, 2023
|
|
8OQ1
 
 | | Crystal structure of tailspike depolymerase (APK14_gp49) from Acinetobacter phage vB_AbaP_APK14 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii.
Int J Mol Sci, 24, 2023
|
|
8PNX
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in PMP form | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of dual substrate recognition in the transaminase from Blastococcus saxobsidens specific to D-amino acids and R-amines
To Be Published
|
|
