1OV7
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with 2-Allyl-6-Methyl-Phenol | | 分子名称: | 2-ALLYL-6-METHYL-PHENOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2003-03-25 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1TLA
 
 | | HYDROPHOBIC CORE REPACKING AND AROMATIC-AROMATIC INTERACTION IN THE THERMOSTABLE MUTANT OF T4 LYSOZYME SER 117 (RIGHT ARROW) PHE | | 分子名称: | CHLORIDE ION, PHOSPHATE ION, T4 LYSOZYME | | 著者 | Anderson, D.E, Hurley, J.H, Nicholson, H, Baase, W.A, Matthews, B.W. | | 登録日 | 1993-03-22 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Hydrophobic core repacking and aromatic-aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117-->Phe.
Protein Sci., 2, 1993
|
|
1OVJ
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with 3-Fluoro-2-Methyl_Aniline | | 分子名称: | 3-FLUORO-2-METHYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2003-03-26 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1PE5
 
 | | Thermolysin with tricyclic inhibitor | | 分子名称: | (6-METHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL)METHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | 著者 | Juers, D, Holland, D, Morgan, B.P, Bartlett, P.A, Matthews, B.W. | | 登録日 | 2003-05-21 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
1PE7
 
 | | Thermolysin with bicyclic inhibitor | | 分子名称: | 2-(4-METHYLPHENOXY)ETHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | 著者 | Juers, D, Yusuff, N, Bartlett, P.A, Matthews, B.W. | | 登録日 | 2003-05-21 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
1PE8
 
 | | Thermolysin with monocyclic inhibitor | | 分子名称: | 2-ETHOXYETHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | 著者 | Juers, D, Pyun, H.-J, Bartlett, P.A, Matthews, B.W. | | 登録日 | 2003-05-21 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
1MAT
 
 | |
1OVH
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 2-Chloro-6-Methyl-Aniline | | 分子名称: | 2-CHLORO-6-METHYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2003-03-26 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OWY
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 2-Propyl-Aniline | | 分子名称: | 2-PROPYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2003-03-31 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1Y3G
 
 | | Crystal Structure of a Silanediol Protease Inhibitor Bound to Thermolysin | | 分子名称: | (2S)-2-{[(AMINOMETHYL)(DIHYDROXY)SILYL]METHYL}-4-METHYLPENTANAL, 3-PHENYLPROPANAL, CALCIUM ION, ... | | 著者 | Juers, D.H, Kim, J, Matthews, B.W, Sieburth, S.M. | | 登録日 | 2004-11-24 | | 公開日 | 2006-01-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Analysis of Silanediols as Transition-State-Analogue Inhibitors of the Benchmark Metalloprotease Thermolysin(,).
Biochemistry, 44, 2005
|
|
1YJ3
 
 | | Crystal structure analysis of product bound methionine aminopeptidase Type 1c from Mycobacterium Tuberculosis | | 分子名称: | BETA-MERCAPTOETHANOL, COBALT (II) ION, METHIONINE, ... | | 著者 | Addlagatta, A, Quillin, M.L, Omotoso, O, Liu, J.O, Matthews, B.W. | | 登録日 | 2005-01-13 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Identification of an SH3-binding motif in a new class of methionine aminopeptidases from Mycobacterium tuberculosis suggests a mode of interaction with the ribosome.
Biochemistry, 44, 2005
|
|
1DYD
 
 | |
1DYF
 
 | |
1DYC
 
 | |
1F4H
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1DYA
 
 | |
1DYG
 
 | |
1DYE
 
 | |
1D3F
 
 | |
1D3M
 
 | | METHIONINE CORE MUTATION | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Matthews, B.W. | | 登録日 | 1999-09-29 | | 公開日 | 1999-09-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Use of differentially substituted selenomethionine proteins in X-ray structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1D2Y
 
 | |
1D2W
 
 | |
1DYB
 
 | |
1DP0
 
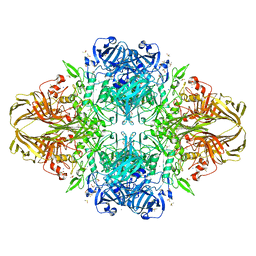 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | 分子名称: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 1999-12-22 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
