2HJI
 
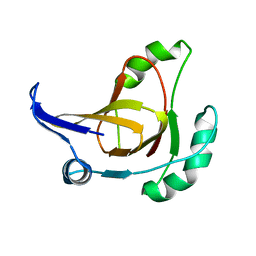 | | Structural model for the Fe-containing isoform of acireductone dioxygenase | | Descriptor: | E-2/E-2' protein, FE (II) ION | | Authors: | Pochapsky, T.C, Ju, T, Maroney, M.J, Chai, S.C. | | Deposit date: | 2006-06-30 | | Release date: | 2006-10-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | One Protein, Two Enzymes Revisited: A Structural Entropy Switch Interconverts the Two Isoforms of Acireductone Dioxygenase
J.Mol.Biol., 363, 2006
|
|
3G4X
 
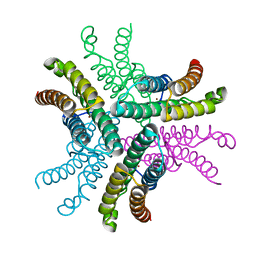 | | Crystal Structure of NiSOD Y9F mutant | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Garman, S.C, Guce, A.I, Herbst, R.W, Bryngelson, P.A, Cabelli, D.E, Higgins, K.A, Ryan, K.C, Maroney, M.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution
Biochemistry, 48, 2009
|
|
3G4Z
 
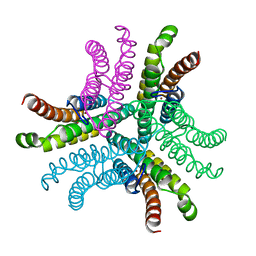 | | Crystal Structure of NiSOD Y9F mutant at 1.9 A | | Descriptor: | BROMIDE ION, NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Garman, S.C, Guce, A.I, Herbst, R.W, Bryngelson, P.A, Cabelli, D.E, Higgins, K.A, Ryan, K.C, Maroney, M.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution
Biochemistry, 48, 2009
|
|
3G50
 
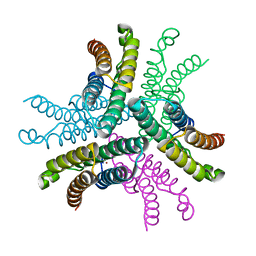 | | Crystal Structure of NiSOD D3A mutant at 1.9 A | | Descriptor: | NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Garman, S.C, Guce, A.I, Herbst, R.W, Bryngelson, P.A, Cabelli, D.E, Higgins, K.A, Ryan, K.C, Maroney, M.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution
Biochemistry, 48, 2009
|
|
4F2E
 
 | |
4NCQ
 
 | | Crystal structure of NiSOD H1A mutant | | Descriptor: | Superoxide dismutase [Ni] | | Authors: | Guce, A.I, Garman, S.C. | | Deposit date: | 2013-10-24 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Nickel superoxide dismutase: structural and functional roles of His1 and its H-bonding network.
Biochemistry, 54, 2015
|
|
3TJA
 
 | | Crystal structure of Helicobacter pylori UreE in the apo form | | Descriptor: | CHLORIDE ION, SULFATE ION, Urease accessory protein ureE | | Authors: | Banaszak, K, Bellucci, M, Zambelli, B, Rypniewski, W.R, Ciurli, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking.
Biochem.J., 441, 2012
|
|
3TJ9
 
 | | Crystal structure of Helicobacter pylori UreE bound to Zn2+ | | Descriptor: | FORMIC ACID, Urease accessory protein ureE, ZINC ION | | Authors: | Banaszak, K, Bellucci, M, Zambelli, B, Rypniewski, W.R, Ciurli, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking.
Biochem.J., 441, 2012
|
|
3TJ8
 
 | | Crystal structure of Helicobacter pylori UreE bound to Ni2+ | | Descriptor: | FORMIC ACID, NICKEL (II) ION, Urease accessory protein ureE | | Authors: | Banaszak, K, Bellucci, M, Zambelli, B, Rypniewski, W.R, Ciurli, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking.
Biochem.J., 441, 2012
|
|
5LBM
 
 | | The asymmetric tetrameric structure of the formaldehyde sensing transcriptional repressor FrmR from Escherichia coli | | Descriptor: | FORMYL GROUP, Transcriptional repressor FrmR | | Authors: | Bisson, C, Baker, P.J, Green, J, Chivers, P.T. | | Deposit date: | 2016-06-16 | | Release date: | 2016-12-21 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of a formaldehyde-sensing transcriptional regulator.
Sci Rep, 6, 2016
|
|
2YJG
 
 | | Structure of the lactate racemase apoprotein from Thermoanaerobacterium thermosaccharolyticum | | Descriptor: | 1,2-ETHANEDIOL, LACTATE RACEMASE APOPROTEIN, MAGNESIUM ION, ... | | Authors: | Declercq, J.P, Desguin, B, Soumillion, P, Hols, P. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactate Racemase is a Nickel-Dependent Enzyme Activated by a Widespread Maturation System.
Nat.Commun., 5, 2014
|
|
4F2F
 
 | | Crystal structure of the metal binding domain (MBD) of the Streptococcus pneumoniae D39 Cu(I) exporting P-type ATPase CopA with Cu(I) | | Descriptor: | CHLORIDE ION, COPPER (I) ION, Cation-transporting ATPase, ... | | Authors: | Fu, Y, Dann III, C.E, Giedroc, D.P. | | Deposit date: | 2012-05-07 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new structural paradigm in copper resistance in Streptococcus pneumoniae.
Nat.Chem.Biol., 9, 2013
|
|
1ZRR
 
 | | Residual Dipolar Coupling Refinement of Acireductone Dioxygenase from Klebsiella | | Descriptor: | E-2/E-2' protein, NICKEL (II) ION | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Ju, T, Hoefler, C, Liang, J. | | Deposit date: | 2005-05-19 | | Release date: | 2005-12-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A refined model for the structure of acireductone dioxygenase from Klebsiella ATCC 8724 incorporating residual dipolar couplings
J.Biomol.NMR, 34, 2006
|
|
