8JZL
 
 | |
8K04
 
 | | CryoEM structure of a 2,3-hydroxycinnamic acid 1,2-dioxygenase MhpB in apo form | | Descriptor: | 2,3-dihydroxyphenylpropionate/2,3-dihydroxicinnamic acid 1,2-dioxygenase | | Authors: | Jiang, W.X, Cheng, X.Q, Ma, L.X, Xing, Q. | | Deposit date: | 2023-07-07 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | CryoEM structure of the NADP-dependent malic enzyme in complex with oxaloacetate
To Be Published
|
|
8K0A
 
 | |
8JZO
 
 | |
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
6J04
 
 | |
7YGI
 
 | | Crystal structure of p53 DBD domain in complex with azurin | | Descriptor: | Azurin, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Jiang, W.X, Zuo, J.Q, Hu, J.J, Chen, X.Q, Ma, L.X, Liu, Z, Xing, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination.
Commun Biol, 6, 2023
|
|
8HKC
 
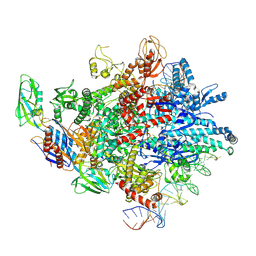 | | Cryo-EM structure of E. coli RNAP sigma32 complex | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wu, S, Ma, L.X. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural Insight into the Mechanism of sigma 32-Mediated Transcription Initiation of Bacterial RNA Polymerase.
Biomolecules, 13, 2023
|
|
