6V7O
 
 | |
7EO0
 
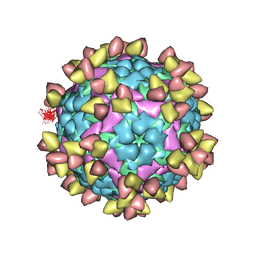 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
7JRA
 
 | |
8GRR
 
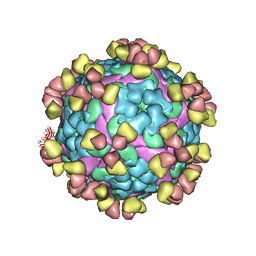 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W125 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Kun, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8GSP
 
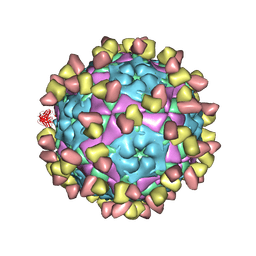 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W2 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
7EL1
 
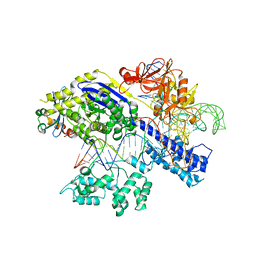 | | Structure of a protein from bacteria | | Descriptor: | 100AA, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Liu, H, Zhu, Y, Huang, Z. | | Deposit date: | 2021-04-07 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis of Staphylococcus aureus Cas9 inhibition by AcrIIA14.
Nucleic Acids Res., 49, 2021
|
|
5VO6
 
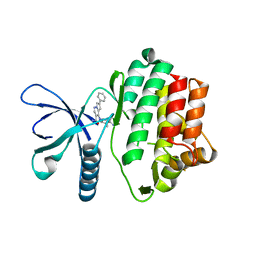 | |
5WFJ
 
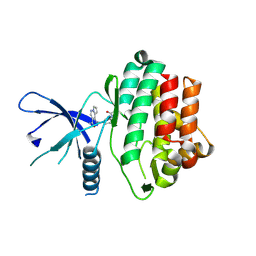 | | THE JAK3 KINASE DOMAIN IN COMPLEX WITH A COVALENT INHIBITOR | | Descriptor: | 4-({[3-(propanoylamino)phenyl]methyl}amino)pyrrolo[1,2-b]pyridazine-3-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Sack, J. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of highly potent, selective, covalent inhibitors of JAK3.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4N75
 
 | |
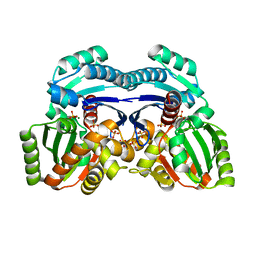
7EPQ
 
 | |
7FAH
 
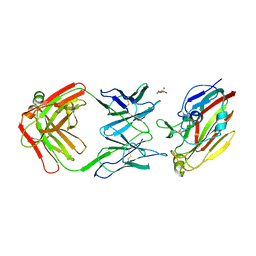 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
4R3H
 
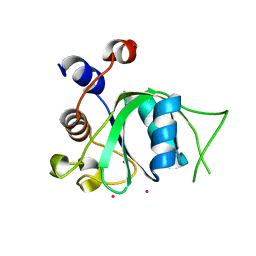 | | The crystal structure of an apo RNA binding protein | | Descriptor: | SULFATE ION, UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Xu, C, Liu, K, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
4R3I
 
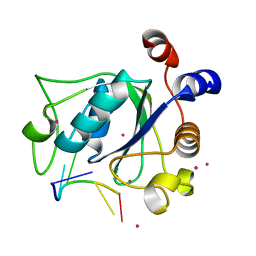 | | The crystal structure of an RNA complex | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Tempel, W, Xu, C, Liu, K, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
4RIO
 
 | |
7ZOP
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with sophorose. | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZON
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with glucose | | Descriptor: | Glycoside hydrolase family 18, PENTAETHYLENE GLYCOL, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOH
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOO
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with gentiobiose | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOI
 
 | | Carbohydrate binding domain CBM92-A from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
6LIT
 
 | | Estrogen-related receptor beta(ERR2) in complex with BPA | | Descriptor: | 10-mer from Nuclear receptor coactivator 2, 4,4'-PROPANE-2,2-DIYLDIPHENOL, Steroid hormone receptor ERR2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-12-13 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Specificity of Ligand Binding and Coactivator Assembly by Estrogen-Related Receptor beta.
J.Mol.Biol., 432, 2020
|
|
6LN4
 
 | | Estrogen-related receptor beta(ERR2) in complex with PGC1a-2a | | Descriptor: | 10-mer from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Steroid hormone receptor ERR2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Insights into the Specificity of Ligand Binding and Coactivator Assembly by Estrogen-Related Receptor beta.
J.Mol.Biol., 432, 2020
|
|
6NX1
 
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN BOUND TETHERED WITH SRC CO-ACTIVATOR PEPTIDE AND COMPOUND-3 AKA 1,1,1,3,3,3-HEXAFLUORO-2-{4-[1-(4- LUOROBENZENESULFONYL)CYCLOPENTYL]PHENYL}PROPAN-2-OL | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-(4-{1-[(4-fluorophenyl)sulfonyl]cyclopentyl}phenyl)propan-2-ol, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 fusion | | Authors: | Khan, J.A. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-based Discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as Selective, Orally Active ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 10, 2019
|
|
6O98
 
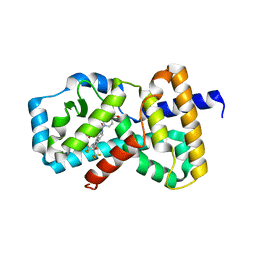 | | Crystal structure of RAR-related orphan receptor C in complex with a phenyl (3-phenylpyrrolidin-3-yl)sulfone inhibitor | | Descriptor: | 1-(4-{(3R)-3-[(4-fluorophenyl)sulfonyl]-3-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]pyrrolidine-1-carbonyl}piperazin-1-yl)ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based Discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as Selective, Orally Active ROR gamma t Inverse Agonists.
ACS Med Chem Lett, 10, 2019
|
|
2TYS
 
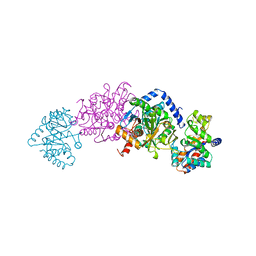 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-L-TRYPTOPHANE | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2TSY
 
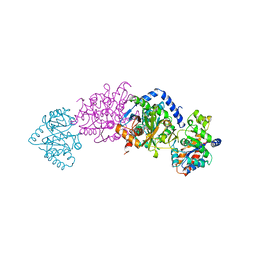 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SN-GLYCEROL-3-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
