3HMC
 
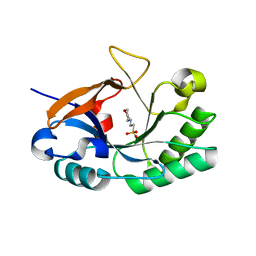 | | Endolysin from Bacillus anthracis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative prophage LambdaBa04, glycosyl hydrolase, ... | | Authors: | Low, L.Y, Liddington, R. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins.
J.Biol.Chem., 286, 2011
|
|
3HMB
 
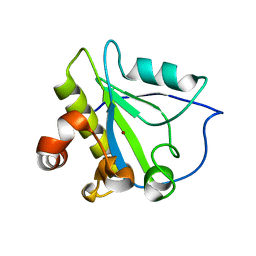 | | Mutant endolysin from Bacillus subtilis | | Descriptor: | N-acetylmuramoyl-L-alanine amidase xlyA, ZINC ION | | Authors: | Low, L.Y, Liddington, R. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins.
J.Biol.Chem., 286, 2011
|
|
3RDR
 
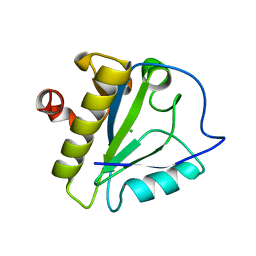 | | Structure of the catalytic domain of XlyA | | Descriptor: | CHLORIDE ION, N-acetylmuramoyl-L-alanine amidase XlyA, ZINC ION | | Authors: | Low, L.Y, Liddington, R.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins.
J.Biol.Chem., 286, 2011
|
|
1YB0
 
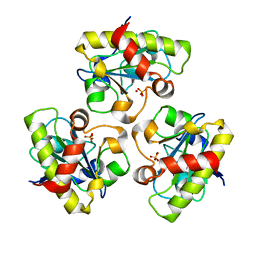 | | Structure of PlyL | | Descriptor: | PHOSPHATE ION, ZINC ION, prophage LambdaBa02, ... | | Authors: | Low, L.Y, Yang, C, Perego, M, Osterman, A, Liddington, R.C. | | Deposit date: | 2004-12-18 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and lytic activity of a Bacillus anthracis prophage endolysin
J.Biol.Chem., 280, 2005
|
|
2AR3
 
 | | E90A mutant structure of PlyL | | Descriptor: | PHOSPHATE ION, ZINC ION, prophage lambdaba02, ... | | Authors: | Low, L.Y, Yang, C, Perego, M, Osterman, A, Liddington, R.C. | | Deposit date: | 2005-08-19 | | Release date: | 2006-06-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and lytic activity of a Bacillus anthracis prophage endolysin.
J.Biol.Chem., 280, 2005
|
|
3JRN
 
 | | Crystal structure of TIR domain from Arabidopsis Thaliana | | Descriptor: | ARSENIC, AT1G72930 protein | | Authors: | Chan, S.L, Mukasa, T, Santelli, E, Low, L.Y, Pascual, J. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants.
Protein Sci., 19, 2009
|
|
8T8S
 
 | | Sortilin-PGRN peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Paragranulin peptide, ... | | Authors: | Srivastava, D.B, Srivastava, A, Cherf, G.M, Low, L.Y, Kannan, G. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural studies of Sortilin-PGRN peptide complex
To Be Published
|
|
8T8R
 
 | | Sortilin-PGRN peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Paragranulin peptide, Sortilin, ... | | Authors: | Srivastava, D.B, Srivastava, A, Cherf, G.M, Low, L.Y, Kannan, G. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural studies of Sortilin-PGRN peptide complex
To Be Published
|
|
1XJH
 
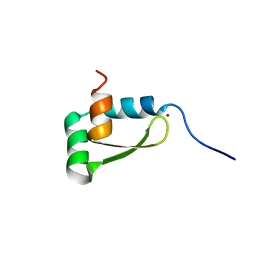 | | NMR structure of the redox switch domain of the E. coli Hsp33 | | Descriptor: | 33 kDa chaperonin, ZINC ION | | Authors: | Won, H.S, Low, L.Y, De Guzman, R.N, Martinez-Yamout, M.A, Jakob, U, Dyson, H.J. | | Deposit date: | 2004-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Zinc-dependent Redox Switch Domain of the Chaperone Hsp33 has a Novel Fold
J.Mol.Biol., 341, 2004
|
|
3H16
 
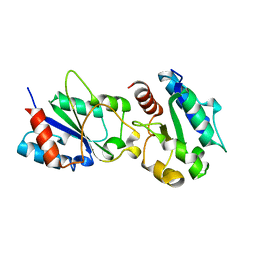 | | Crystal structure of a bacteria TIR domain, PdTIR from Paracoccus denitrificans | | Descriptor: | SULFATE ION, TIR protein | | Authors: | Chan, S.L, Low, L.Y, Santelli, E, Pascual, J. | | Deposit date: | 2009-04-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mimicry in Innate Immunity: CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN.
J.Biol.Chem., 284, 2009
|
|
