7T42
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 2c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T44
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 4c | | Descriptor: | (1R,2S)-2-[(N-{[(2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[2-(methanesulfonyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7TQ8
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 14d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, Orf1a protein, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
3Q8A
 
 | | Crystal structure of WT Protective Antigen (pH 5.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Rajapaksha, M, Lovell, S, Janowiak, B.E, Andra, K.K, Battaile, K.P, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q7R
 
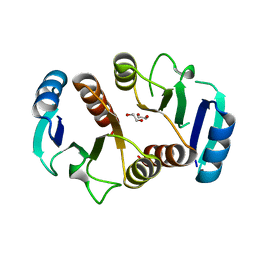 | | 1.6A resolution structure of the ChxR receiver domain from Chlamydia trachomatis | | Descriptor: | 1,2-ETHANEDIOL, Transcriptional regulatory protein | | Authors: | Hickey, J, Lovell, S, Battaile, K.P, Hu, L, Middaugh, C.R, Hefty, P.S. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.
J.Biol.Chem., 286, 2011
|
|
3QH6
 
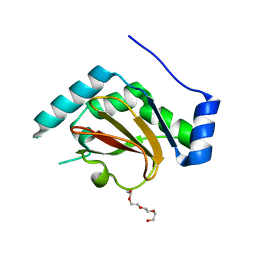 | | 1.8A resolution structure of CT296 from Chlamydia trachomatis | | Descriptor: | CT296, TETRAETHYLENE GLYCOL | | Authors: | Kemege, K, Hickey, J, Lovell, S, Battaile, K.P, Zhang, Y, Hefty, P.S. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes.
J.Bacteriol., 193, 2011
|
|
3Q7T
 
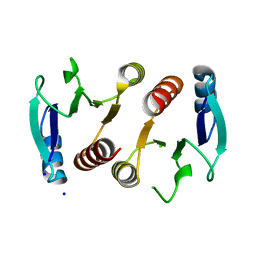 | | 2.15A resolution structure (I41 Form) of the ChxR receiver domain from Chlamydia trachomatis | | Descriptor: | SODIUM ION, Transcriptional regulatory protein | | Authors: | Hickey, J, Lovell, S, Battaile, K.P, Hu, L, Middaugh, C.R, Hefty, P.S. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.
J.Biol.Chem., 286, 2011
|
|
3Q7S
 
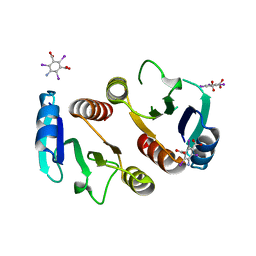 | | 2.1A resolution structure of the ChxR receiver domain containing I3C from Chlamydia trachomatis | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Transcriptional regulatory protein | | Authors: | Hickey, J, Lovell, S, Battaile, K.P, Hu, L, Middaugh, C.R, Hefty, P.S. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.
J.Biol.Chem., 286, 2011
|
|
3QH7
 
 | | 2.5 A resolution structure of Se-Met labeled CT296 from Chlamydia trachomatis | | Descriptor: | CT296 | | Authors: | Kemege, K, Hickey, J, Lovell, S, Battaile, K.P, Zhang, Y, Hefty, P.S. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-05 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes.
J. Bacteriol., 193, 2011
|
|
8DOP
 
 | |
8DOS
 
 | |
8DOR
 
 | |
8EK7
 
 | |
8U91
 
 | |
8U8W
 
 | |
8U90
 
 | |
8U93
 
 | |
8U92
 
 | |
8UR1
 
 | |
8SLD
 
 | |
8SLF
 
 | |
8SLH
 
 | |
8T1A
 
 | |
8FI5
 
 | |
8FI6
 
 | |
