6KLM
 
 | | NMR solution structure of Roseltide rT7 | | Descriptor: | Roseltide rT7 | | Authors: | Fan, J.S, Kam, A, Loo, S, Yang, D, Tam, P.J. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Roseltide rT7 is a disulfide-rich, anionic, and cell-penetrating peptide that inhibits proteasomal degradation.
J.Biol.Chem., 294, 2019
|
|
2HTN
 
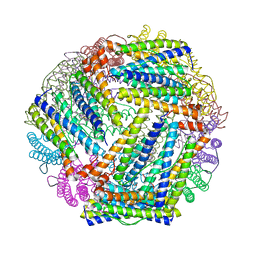 | | E. coli bacterioferritin in its as-isolated form | | Descriptor: | Bacterioferritin, FE (III) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | van Eerde, A, Wolterink-Van Loo, S, Van Der Oost, J, Dijkstra, B.W. | | Deposit date: | 2006-07-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fortuitous structure determination of 'as-isolated' Escherichia coli bacterioferritin in a novel crystal form.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
7V5E
 
 | |
7V5F
 
 | |
5GSF
 
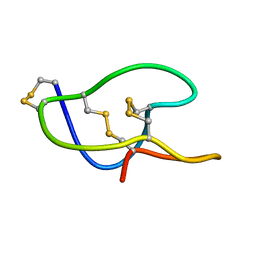 | | Structure of roseltide rT1 | | Descriptor: | roseltide rT1 | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of Roseltide, a Knottin-type Neutrophil Elastase Inhibitor Derived from Hibiscus sabdariffa.
Sci Rep, 6, 2016
|
|
5XBD
 
 | |
2NUY
 
 | |
2NUX
 
 | |
2NUW
 
 | |
