2GJI
 
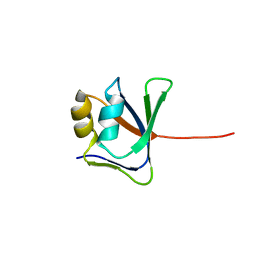 | | NMR solution structure of VP9 from White Spot Syndrome Virus | | Descriptor: | wsv230 | | Authors: | Liu, Y, Wu, J.L, Song, J.X, Sivaraman, J, Hew, C.L. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Identification of a Novel Nonstructural Protein, VP9, from White Spot Syndrome Virus: Its Structure Reveals a Ferredoxin Fold with Specific Metal Binding Sites
J.Virol., 80, 2006
|
|
4O6X
 
 | |
5T1G
 
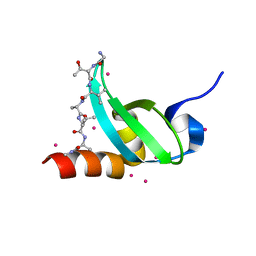 | | chromo shadow domain of CBX1 in complex with a histone peptide | | Descriptor: | Chromobox protein homolog 1, Histone H3.1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | chromo shadow domain of CBX1 in complex with a histone peptide
To Be Published
|
|
5UK2
 
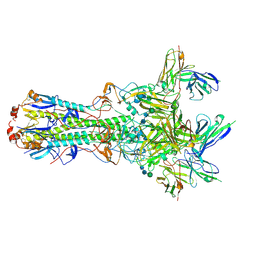 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UJZ
 
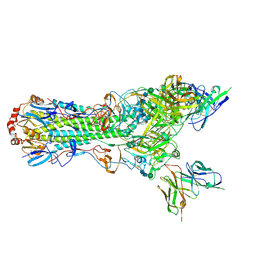 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
6JPI
 
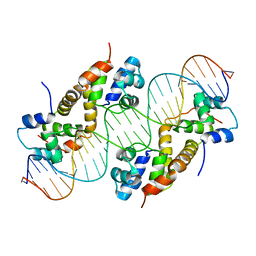 | |
4Q46
 
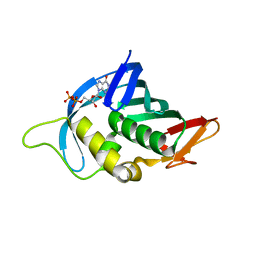 | |
5UK0
 
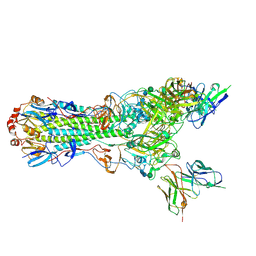 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UZA
 
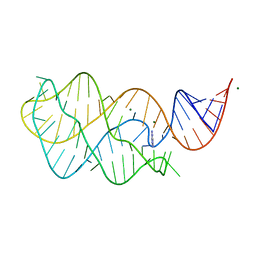 | | Adenine riboswitch aptamer domain labelled with iodo-uridine by position-selective labelling of RNA (PLOR) | | Descriptor: | ADENINE, MAGNESIUM ION, RNA (71-MER) | | Authors: | Liu, Y, Stagno, J.R, Wang, Y.-X. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA.
Nat Protoc, 13, 2018
|
|
5UK1
 
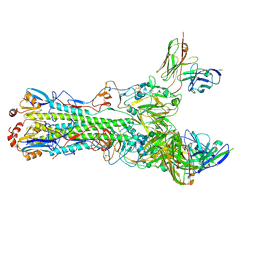 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
4GZN
 
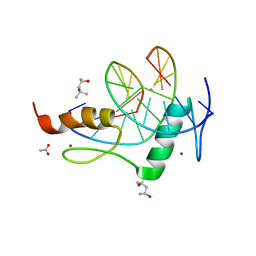 | | Mouse ZFP57 zinc fingers in complex with methylated DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liu, Y, Zhang, X, Cheng, X. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence.
Genes Dev., 26, 2012
|
|
4OR6
 
 | |
4OR4
 
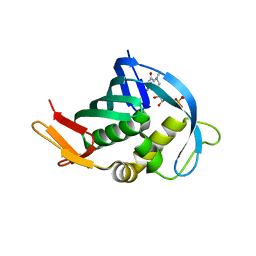 | |
3BGJ
 
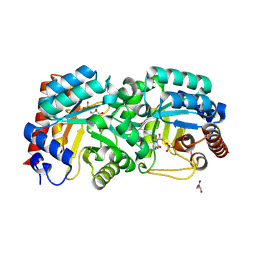 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 6-iodo-UMP | | Descriptor: | GLYCEROL, URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Bello, A.M, Devalla, S, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 6-iodo-UMP
To be Published
|
|
3BGG
 
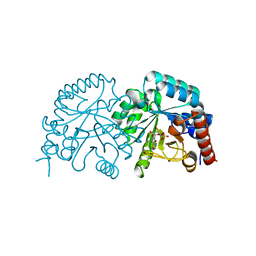 | | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Wang, X.Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP
To be Published
|
|
4QQ6
 
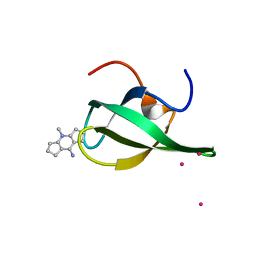 | | Crystal Structure of tudor domain of SMN1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
4QQD
 
 | | Crystal Structure of tandem tudor domains of UHRF1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, E3 ubiquitin-protein ligase UHRF1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
4RZH
 
 | | Crystal structure of FabG from Synechocystis sp. PCC 6803 | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Liu, Y, Xue, S. | | Deposit date: | 2014-12-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-directed construction of a high-performance version of the enzyme FabG from the photosynthetic microorganism Synechocystis sp. PCC 6803.
Febs Lett., 589, 2015
|
|
3BPW
 
 | |
3BK0
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Complexed with 5-CN-UMP | | Descriptor: | 5-CYANO-URIDINE-5'-MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, Y, Tang, H.L, Avina, M.E.M, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-12-05 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Complexed with 5-CN-UMP
To be Published
|
|
3BVJ
 
 | |
3PST
 
 | |
3PSP
 
 | |
3Q9S
 
 | |
3Q9V
 
 | |
