4WCJ
 
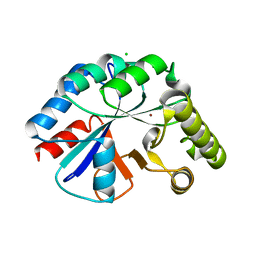 | | Structure of IcaB from Ammonifex degensii | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
5WEZ
 
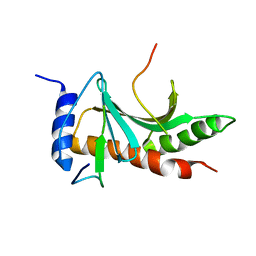 | | Structure of the Tir-CesT effector-chaperone complex | | Descriptor: | Tir chaperone, Translocated intimin receptor Tir | | Authors: | Little, D.J, Coombes, B.K. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular basis for CesT recognition of type III secretion effectors in enteropathogenic Escherichia coli.
PLoS Pathog., 14, 2018
|
|
5BU6
 
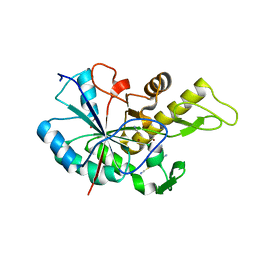 | | Structure of BpsB deaceylase domain from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, BpsB (PgaB), Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Little, D.J, Bamford, N.C, Howell, P.L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The Protein BpsB Is a Poly-beta-1,6-N-acetyl-d-glucosamine Deacetylase Required for Biofilm Formation in Bordetella bronchiseptica.
J.Biol.Chem., 290, 2015
|
|
4F9D
 
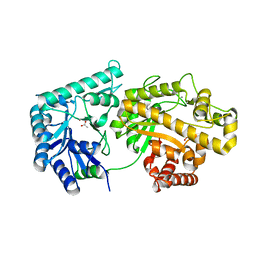 | | Structure of Escherichia coli PgaB 42-655 in complex with nickel | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
4F9J
 
 | | Structure of Escherichia coli PgaB 42-655 in complex with iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
6AU1
 
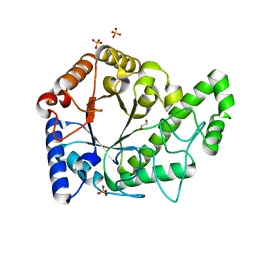 | | Structure of the PgaB (BpsB) glycoside hydrolase domain from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative hemin storage protein, ... | | Authors: | Little, D.J, Bamford, N.C, Howell, P.L. | | Deposit date: | 2017-08-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-beta (1,6)-N-acetylglucosamine and can disrupt bacterial biofilms.
PLoS Pathog., 14, 2018
|
|
4P7L
 
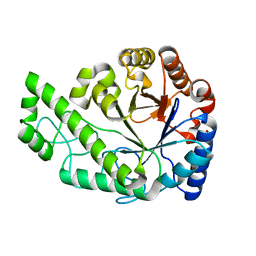 | | Structure of Escherichia coli PgaB C-terminal domain, P212121 crystal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7O
 
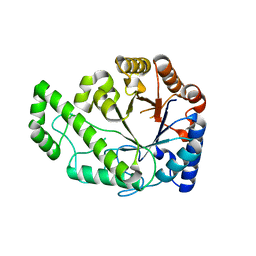 | | Structure of Escherichia coli PgaB C-terminal domain, P1 crystal form | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7Q
 
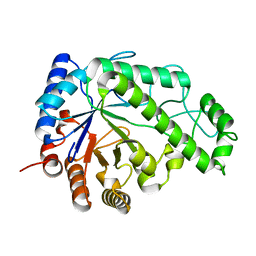 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7R
 
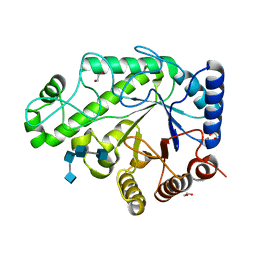 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7N
 
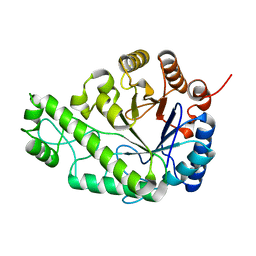 | | Structure of Escherichia coli PgaB C-terminal domain in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5BX9
 
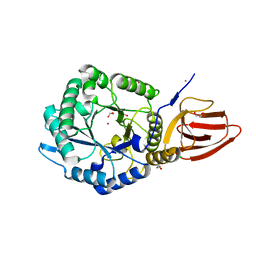 | | Structure of PslG from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Baker, P, Little, D.J, Howell, P.L. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa Glycoside Hydrolase PslG Reveals That Its Levels Are Critical for Psl Polysaccharide Biosynthesis and Biofilm Formation.
J.Biol.Chem., 290, 2015
|
|
5BXA
 
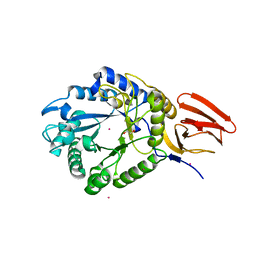 | | Structure of PslG from Pseudomonas aeruginosa in complex with mannose | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Baker, P, Little, D.J, Howell, P.L. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa Glycoside Hydrolase PslG Reveals That Its Levels Are Critical for Psl Polysaccharide Biosynthesis and Biofilm Formation.
J.Biol.Chem., 290, 2015
|
|
7T8N
 
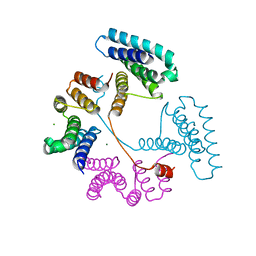 | | Crystal structure of the PNAG binding module PgaA-TPR 220-359 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Poly-beta-1,6-N-acetyl-D-glucosamine export protein | | Authors: | Pfoh, R, Little, D.J, Howell, P.L. | | Deposit date: | 2021-12-16 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The TPR domain of PgaA is a multifunctional scaffold that binds PNAG and modulates PgaB-dependent polymer processing.
Plos Pathog., 18, 2022
|
|
5C5G
 
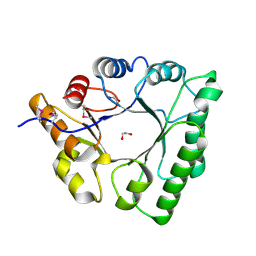 | |
5D6T
 
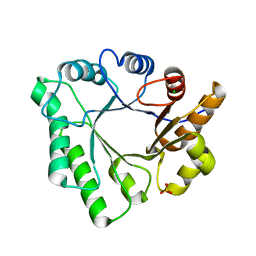 | | Crystal Structure of Aspergillus clavatus Sph3 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, SPHERULIN-4, ... | | Authors: | Bamford, N.C, Little, D.J, Howell, P.L. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Sph3 Is a Glycoside Hydrolase Required for the Biosynthesis of Galactosaminogalactan in Aspergillus fumigatus.
J.Biol.Chem., 290, 2015
|
|
5UG1
 
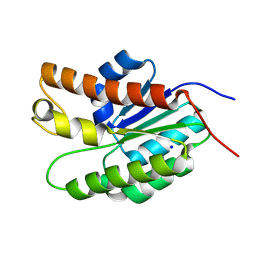 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain with methylsulfonyl adduct | | Descriptor: | Acyltransferase, SODIUM ION, methanesulfonic acid | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5UFY
 
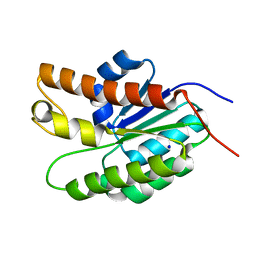 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acyltransferase, SODIUM ION | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5V8E
 
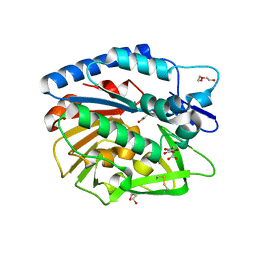 | | Structure of Bacillus cereus PatB1 | | Descriptor: | Bacillus cereus PatB1, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
5V8D
 
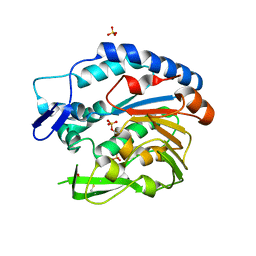 | | Structure of Bacillus cereus PatB1 with sulfonyl adduct | | Descriptor: | Bacillus cereus PatB1, SULFATE ION | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
4O8V
 
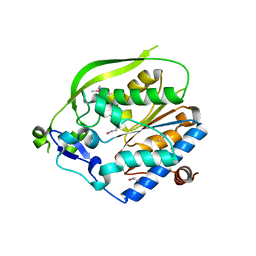 | | O-Acetyltransferase Domain of Pseudomonas putida AlgJ | | Descriptor: | Alginate biosynthesis protein AlgJ | | Authors: | Ricer, T, Little, D.J, Whitney, J.C, Robinson, H, Howell, P.L. | | Deposit date: | 2013-12-30 | | Release date: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | P. aeruginosa SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation.
Plos Pathog., 10, 2014
|
|
5EQ6
 
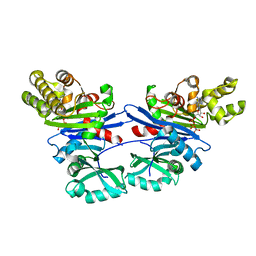 | | Pseudomonas aeruginosa PilM bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EOY
 
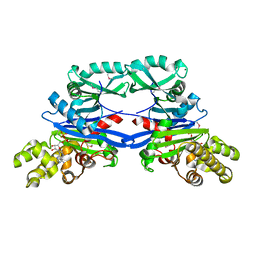 | | Pseudomonas aeruginosa SeMet-PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EOU
 
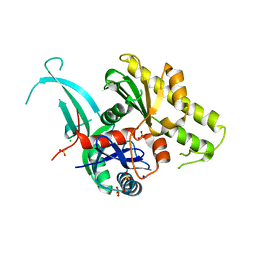 | | Pseudomonas aeruginosa PilM:PilN1-12 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L, Howell, L.P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EOX
 
 | | Pseudomonas aeruginosa PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
