1Z2T
 
 | | NMR structure study of anchor peptide Ser65-Leu87 of enzyme acholeplasma laidlawii Monoglycosyldiacyl Glycerol Synthase (alMGS) in DHPC micelles | | 分子名称: | Anchor peptide Ser65-Leu87 of alMGS | | 著者 | Lind, J, Barany-Wallje, E, Ramo, T, Wieslander, A, Maler, L. | | 登録日 | 2005-03-09 | | 公開日 | 2006-03-21 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, position of and membrane-interaction of a putative membrane-anchoring domain of alMGS
To be Published
|
|
1MVR
 
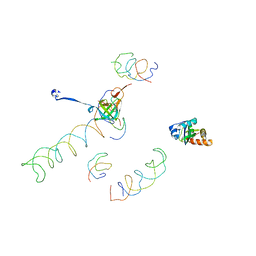 | | Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome | | 分子名称: | 30S RIBOSOMAL PROTEIN S12, 50S ribosomal protein L11, Helix 34 of 16S rRNA, ... | | 著者 | Rawat, U.B, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | 登録日 | 2002-09-26 | | 公開日 | 2003-04-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12.8 Å) | | 主引用文献 | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
2L7C
 
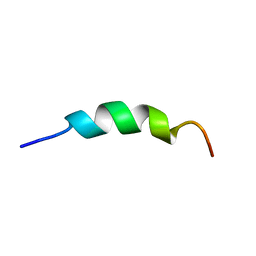 | | Biophysical studies of lipid interacting regions of DGD2 in Arabidopsis thaliana | | 分子名称: | Digalactosyldiacylglycerol synthase 2, chloroplastic | | 著者 | Szpryngiel, S, Ge, C, Iakovleva, I, Lind, J, Wieslander, A, Maler, L. | | 登録日 | 2010-12-07 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Lipid interacting regions in phosphate stress glycosyltransferase atDGD2 from Arabidopsis thaliana.
Biochemistry, 50, 2011
|
|
1JQM
 
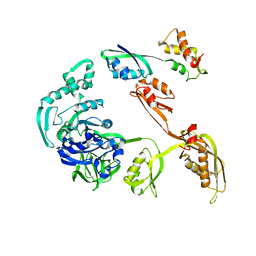 | | Fitting of L11 protein and elongation factor G (EF-G) in the cryo-em map of e. coli 70S ribosome bound with EF-G, GDP and fusidic acid | | 分子名称: | 50S Ribosomal protein L11, Elongation Factor G | | 著者 | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | 登録日 | 2001-08-07 | | 公開日 | 2001-09-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQS
 
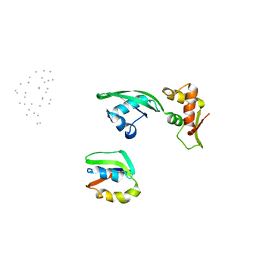 | | Fitting of L11 protein and elongation factor G (domain G' and V) in the cryo-em map of E. coli 70S ribosome bound with EF-G and GMPPCP, a nonhydrolysable GTP analog | | 分子名称: | 50S Ribosomal protein L11, Elongation Factor G | | 著者 | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | 登録日 | 2001-08-07 | | 公開日 | 2001-09-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQT
 
 | | Fitting of L11 protein in the low resolution cryo-EM map of E.coli 70S ribosome | | 分子名称: | 50S Ribosomal protein L11 | | 著者 | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | 登録日 | 2001-08-07 | | 公開日 | 2001-09-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
2K44
 
 | |
1MI6
 
 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | 分子名称: | peptide chain release factor RF-2 | | 著者 | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | 登録日 | 2002-08-22 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12.8 Å) | | 主引用文献 | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
2KJW
 
 | |
2KJV
 
 | |
