5ZUB
 
 | |
5ZU9
 
 | |
5ZU7
 
 | |
5ZUA
 
 | |
5DUS
 
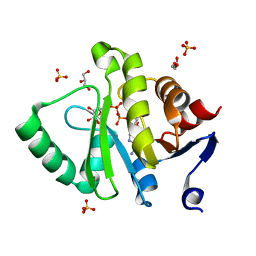 | | Crystal structure of MERS-CoV macro domain in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, ORF1a, ... | | Authors: | Cho, C.-C, Lin, M.-H, Chuang, C.-Y, Hsu, C.-H. | | Deposit date: | 2015-09-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Macro Domain from Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Is an Efficient ADP-ribose Binding Module: CRYSTAL STRUCTURE AND BIOCHEMICAL STUDIES
J.Biol.Chem., 291, 2016
|
|
3AHY
 
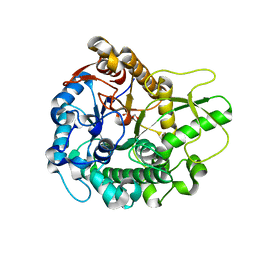 | | Crystal structure of beta-glucosidase 2 from fungus Trichoderma reesei in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase | | Authors: | Jeng, W.-Y, Liu, C.-I, Wang, A.H.-J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis
J.Struct.Biol., 173, 2011
|
|
3AHZ
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, GLYCEROL | | Authors: | Jeng, W.-Y, Liu, C.-I, Wang, A.H.-J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis
J.Struct.Biol., 173, 2011
|
|
3AHX
 
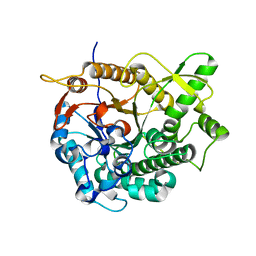 | |
3AI0
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with para-nitrophenyl-beta-D-glucopyranoside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranoside, GLYCEROL, beta-glucosidase | | Authors: | Jeng, W.-Y, Liu, C.-I, Wang, A.H.-J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis
J.Struct.Biol., 173, 2011
|
|
