4R99
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | SULFATE ION, Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
8YQF
 
 | |
8W9L
 
 | |
8W9X
 
 | |
8WLD
 
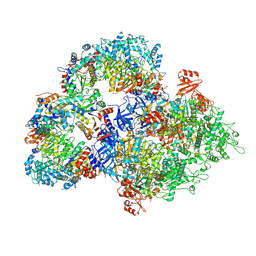 | | Cryo-EM structure of SIR2/HerA antiphage complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Cryo-EM structure of SIR2/HerA antiphage complex
To Be Published
|
|
8WIV
 
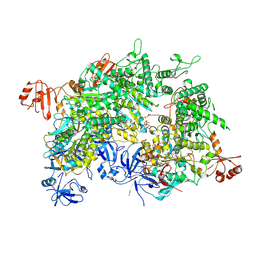 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WOC
 
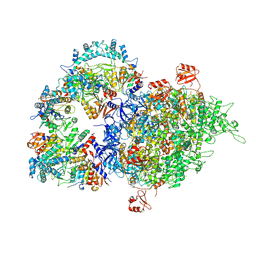 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WOF
 
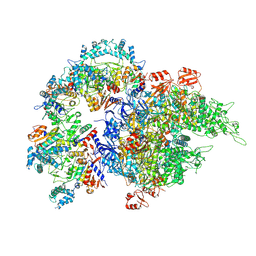 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WJ3
 
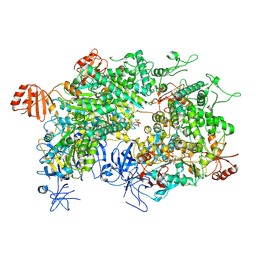 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WK0
 
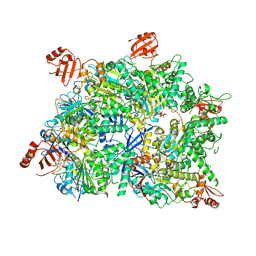 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WOD
 
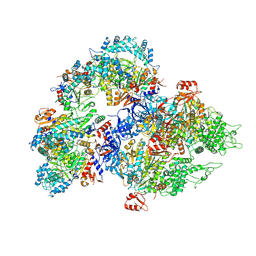 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
5HWL
 
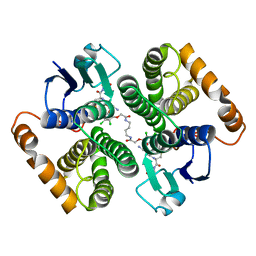 | | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 2, N,N'-(butane-1,4-diyl)bis{2-[2,3-dichloro-4-(2-methylidenebutanoyl)phenoxy]acetamide} | | Authors: | Zhang, X, Wei, J, Wu, S, Zhang, H.P, Luo, M, Yang, X.L, Liao, F, Wang, D.Q. | | Deposit date: | 2016-01-29 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form
To Be Published
|
|
5B85
 
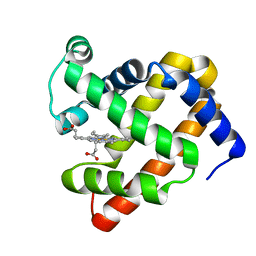 | | X-ray structure of ferric F138Y sperm whale myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuan, H. | | Deposit date: | 2016-06-12 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distinct roles of a tyrosine-associated hydrogen-bond network in fine-tuning the structure and function of heme proteins: two cases designed for myoglobin
Mol Biosyst, 12, 2016
|
|
5B84
 
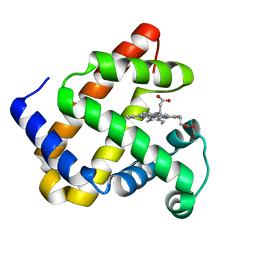 | | X-ray crystal structure of met I107Y sperm whale myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuan, H. | | Deposit date: | 2016-06-12 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Distinct roles of a tyrosine-associated hydrogen-bond network in fine-tuning the structure and function of heme proteins: two cases designed for myoglobin
Mol Biosyst, 12, 2016
|
|
5XKV
 
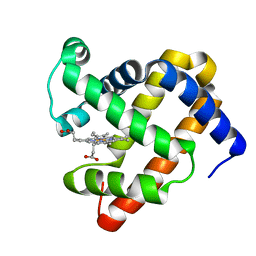 | | structure of sperm whale myoglobin F138W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuan, H. | | Deposit date: | 2017-05-09 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational design of artificial dye-decolorizing peroxidases using myoglobin by engineering Tyr/Trp in the heme center
Dalton Trans, 46, 2017
|
|
5XKW
 
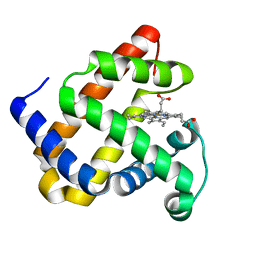 | | myoglobin mutant F43Y/F46Y | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuan, H. | | Deposit date: | 2017-05-09 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational design of artificial dye-decolorizing peroxidases using myoglobin by engineering Tyr/Trp in the heme center
Dalton Trans, 46, 2017
|
|
8GNA
 
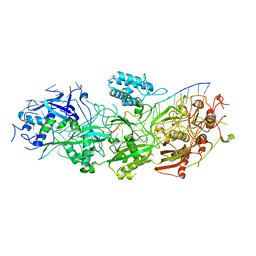 | | Structure of the SbCas7-11-crRNA-NTR complex | | Descriptor: | RAMP superfamily protein, RNA (32-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
8YHE
 
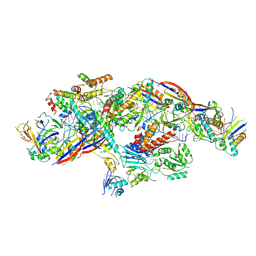 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state II | | Descriptor: | RNA (29-MER), RNA (46-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z9C
 
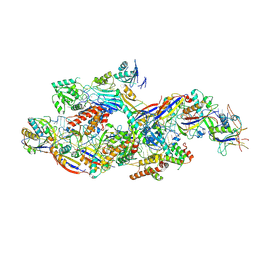 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | Protein structure, RNA (41-MER), RNA (48-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4L
 
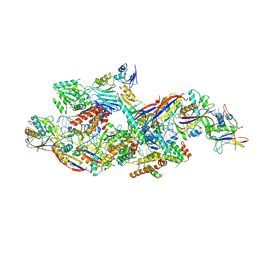 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | RNA (40-MER), RNA (49-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z9E
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (39-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4J
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (38-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z99
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state +I | | Descriptor: | RNA (49-MER), RNA (54-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8YHD
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state I | | Descriptor: | RNA (35-MER), RNA (53-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
