5ZMZ
 
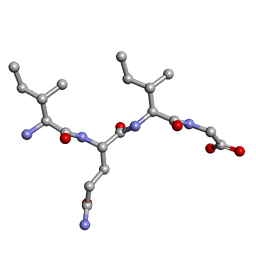 | | Amyloid core of RIP1 | | Descriptor: | Amyloid core of RIP1 | | Authors: | Li, J.X, Zheng, J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Amyloid core of RIP1
To Be Published
|
|
8XAT
 
 | |
8XAS
 
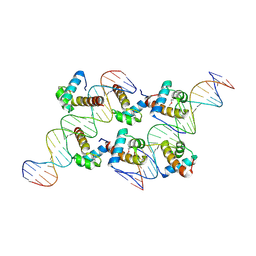 | | Crystal structure of AtARR1-DBD in complex with a DNA fragment | | Descriptor: | DNA (50-MER), Two-component response regulator ARR1 | | Authors: | Li, J.X, Zhou, C.M, zhang, P, Wang, J.W. | | Deposit date: | 2023-12-05 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5Z87
 
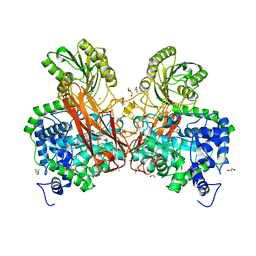 | | Structural of a novel b-glucosidase EmGH1 at 2.3 angstrom from Erythrobacter marinus | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BENZAMIDINE, ... | | Authors: | Li, J.X, Hu, X.J, Zhao, Y, Li, L. | | Deposit date: | 2018-01-31 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analysis of a novel b-glucosidase EmGH1 from Erythrobacter marinus
To Be Published
|
|
7YJ9
 
 | |
7YJA
 
 | |
7YJB
 
 | | Crystal structure of Stenoloma chusanum chalcone synthase 1 (ScCHS1) complex with CoA and Eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, chalcone synthase 1 (ScCHS1) | | Authors: | Li, J.X, Cheng, A.X, Zhang, P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and structural characterization of a promiscuous chalcone synthase from the fern species Stenoloma chusanum.
J Integr Plant Biol, 64, 2022
|
|
7YJ6
 
 | |
7YJ8
 
 | |
7YJ5
 
 | |
7YJ7
 
 | |
5YZL
 
 | |
5GSK
 
 | | Crystal structure of duplex DNA3 in complex with Hg(II) and Sr(II) | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), MERCURY (II) ION, STRONTIUM ION | | Authors: | Liu, H.H, Wang, R, Yao, Q.Q, Cheng, Y.Q, Yang, C, Luo, Q, Wu, B.X, Li, J.X, Ma, J.B, Sheng, J, Gan, J.H. | | Deposit date: | 2016-08-16 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
6KEU
 
 | | Wildtype E53, a microbial HSL esterase | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Yang, X.C, Li, Z.Y, Xu, X.W, Li, J.X. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Wildtype E53, a microbial HSL esterase
To Be Published
|
|
5YZJ
 
 | |
7C5F
 
 | | Crystal Structure of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.88 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Characterization and structure of glyceraldehyde-3-phosphate dehydrogenase type 1 from Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4GAX
 
 | | Crystal Structure of an alpha-Bisabolol synthase mutant | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Li, J, Peng, Z. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
4FJQ
 
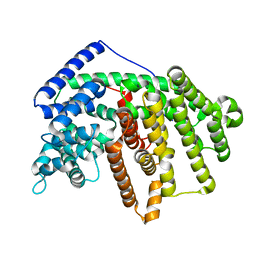 | | Crystal Structure of an alpha-Bisabolol synthase | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Jianxu, L, Peng, Z. | | Deposit date: | 2012-06-12 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0001 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
7BZB
 
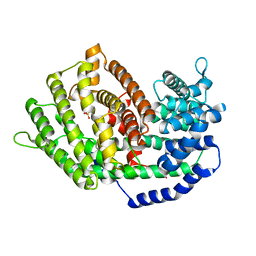 | |
7BZC
 
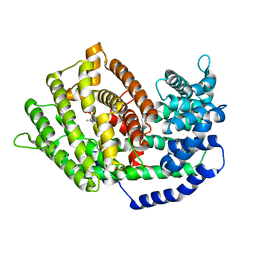 | | Crystal structure of plant sesterterpene synthase AtTPS18 complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, Terpenoid synthase 18 | | Authors: | Li, J.X, Wang, G.D, Zhang, P. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Molecular Basis for Sesterterpene Diversity Produced by Plant Terpene Synthases.
Plant Commun., 1, 2020
|
|
6KVL
 
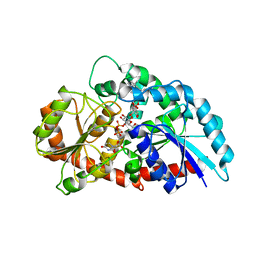 | | Crystal structure of UDP-RebB-SrUGT76G1 | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVI
 
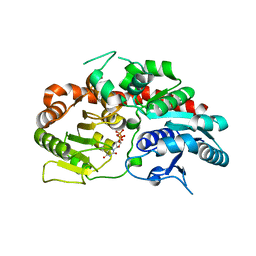 | | Crystal structure of UDP-SrUGT76G1 | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVJ
 
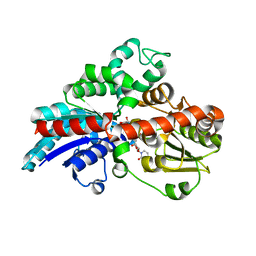 | | Crystal structure of UDPX-SrUGT76G1 | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE-XYLOPYRANOSE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVK
 
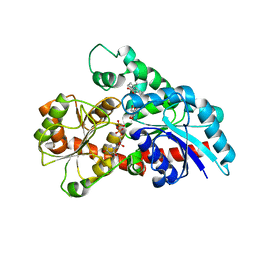 | | Crystal structure of UDP-Sm-SrUGT76G1 | | Descriptor: | Steviolmonoside, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KZ8
 
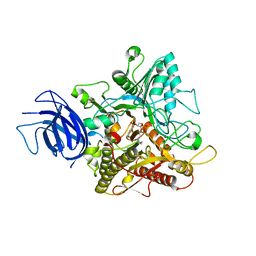 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
