1TZI
 
 | |
1YCS
 
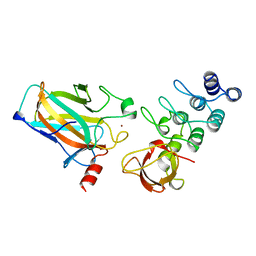 | | P53-53BP2 COMPLEX | | Descriptor: | 53BP2, P53, ZINC ION | | Authors: | Gorina, S, Pavletich, N.P. | | Deposit date: | 1996-09-30 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2.
Science, 274, 1996
|
|
7JVF
 
 | |
7JU9
 
 | |
