3USY
 
 | |
4FQ0
 
 | |
4GC8
 
 | |
3GWG
 
 | |
3H1G
 
 | |
3H1F
 
 | |
3H1E
 
 | | Crystal structure of Mg(2+) and BeH(3)(-)-bound CheY of Helicobacter pylori | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY homolog, MAGNESIUM ION, ... | | Authors: | Lam, K.H, Ling, T.K, Au, S.W. | | Deposit date: | 2009-04-12 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of activated CheY1 from Helicobacter pylori.
J.Bacteriol., 192, 2010
|
|
3USW
 
 | |
5XRW
 
 | |
8FBF
 
 | | Crystal structure of OrfX2 from Clostridium botulinum E1 | | Descriptor: | Neurotoxin complex component Orf-X2, SULFATE ION | | Authors: | Lam, K.H, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
2G4D
 
 | | Crystal structure of human SENP1 mutant (C603S) in complex with SUMO-1 | | Descriptor: | SENP1 protein, Small ubiquitin-related modifier 1 | | Authors: | Xu, Z, Chau, S.F, Lam, K.H, Au, S.W.N. | | Deposit date: | 2006-02-22 | | Release date: | 2006-10-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease
Biochem.J., 398, 2006
|
|
5WUJ
 
 | | Crystal structure of FliF-FliG complex from H. pylori | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, GLYCEROL | | Authors: | Au, S.W, Xue, C, Lam, K.H, Lee, S.H. | | Deposit date: | 2016-12-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum
J. Biol. Chem., 293, 2018
|
|
7M1H
 
 | |
7LZP
 
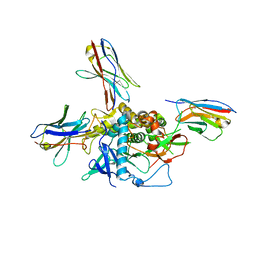 | | LC/A-JPU-B9-JPU-A11-JPU-G11 | | Descriptor: | Botulinum neurotoxin A light chain, JPU-A11, JPU-B9, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2021-03-10 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Probing the structure and function of the protease domain of botulinum neurotoxins using single-domain antibodies.
Plos Pathog., 18, 2022
|
|
7NA9
 
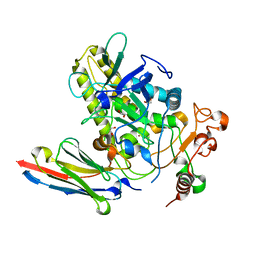 | | Crystal structure of BoNT/B-LC-JSG-C1 | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type B, JSG-C1, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Probing the structure and function of the protease domain of botulinum neurotoxins using single-domain antibodies.
Plos Pathog., 18, 2022
|
|
6BVD
 
 | | Structure of Botulinum Neurotoxin Serotype HA Light Chain | | Descriptor: | ACETATE ION, CALCIUM ION, Light Chain, ... | | Authors: | Jin, R, Lam, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical characterization of the protease domain of the mosaic botulinum neurotoxin type HA.
Pathog Dis, 76, 2018
|
|
6DKK
 
 | | Structure of BoNT | | Descriptor: | Botulinum neurotoxin type A, PHOSPHATE ION | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1.
Nat Commun, 9, 2018
|
|
6MHJ
 
 | | Structure of BoNT mutant | | Descriptor: | Botulinum neurotoxin type A, PHOSPHATE ION | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2018-09-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.019 Å) | | Cite: | A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1.
Nat Commun, 9, 2018
|
|
7T5F
 
 | |
7K7Y
 
 | |
7K84
 
 | |
7L6V
 
 | |
5WIX
 
 | | Crystal structure of P47 of Clostridium botulinum E1 | | Descriptor: | p-47 protein | | Authors: | Lam, K, Liu, S, Qi, R, Jin, R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The hypothetical protein P47 of Clostridium botulinum E1 strain Beluga has a structural topology similar to bactericidal/permeability-increasing protein.
Toxicon, 147, 2018
|
|
6C0B
 
 | | Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-2, MALONATE ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2017-12-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of frizzled proteins byClostridium difficiletoxin B.
Science, 360, 2018
|
|
6OQ7
 
 | | Structure of the GTD domain of Clostridium difficile toxin B in complex with VHH E3 | | Descriptor: | E3, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
