8DMO
 
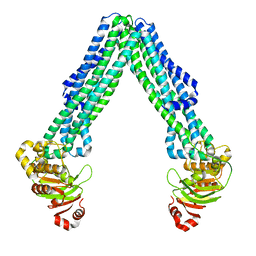 | | Structure of open, inward-facing MsbA from E. coli | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Liu, C, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2022-07-08 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for lipid and copper regulation of the ABC transporter MsbA.
Nat Commun, 13, 2022
|
|
8DMM
 
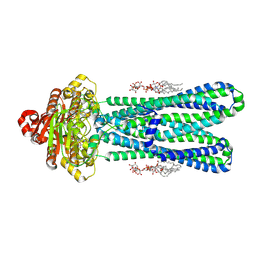 | | Structure of the vanadate-trapped MsbA bound to KDL | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ADP ORTHOVANADATE, ATP-binding transport protein MsbA | | Authors: | Liu, C, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2022-07-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for lipid and copper regulation of the ABC transporter MsbA.
Nat Commun, 13, 2022
|
|
8TSR
 
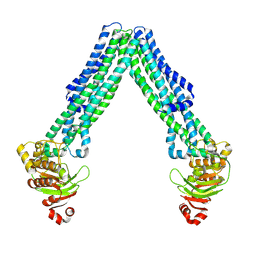 | | Open, inward-facing MsbA structure (OIF4) | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
8TSP
 
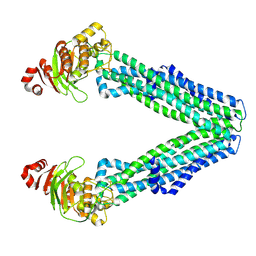 | | Open, inward-facing MsbA structure (OIF1) | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
8TSO
 
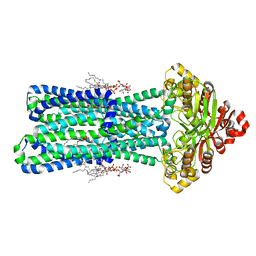 | | KDL bound, nucleotide-free MsbA in open, outward-facing conformation | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-binding transport protein MsbA, PENTAETHYLENE GLYCOL MONODECYL ETHER | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
8TSQ
 
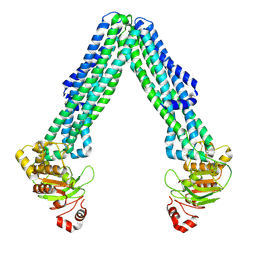 | | Open, inward-facing MsbA structure (OIF2) | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
8TSS
 
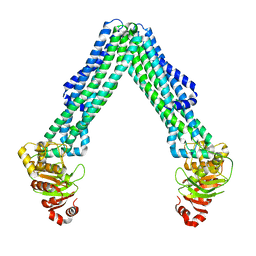 | | Open, inward-facing MsbA structure (OIF3) | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2023-08-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native mass spectrometry and structural studies reveal modulation of MsbA-nucleotide interactions by lipids.
Nat Commun, 15, 2024
|
|
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX4
 
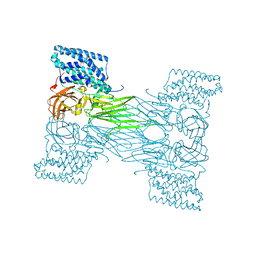 | | mosquitocidal Cry11Aa determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX6
 
 | | mosquitocidal Cry11Aa-E583Q determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX5
 
 | | mosquitocidal Cry11Aa-Y449F determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX7
 
 | | mosquitocidal Cry11Aa-F17Y determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7R1E
 
 | | Mosquitocidal Cry11Ba determined at pH 10.4 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | GLYCEROL, Pesticidal crystal protein Cry11Ba | | Authors: | Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
8ELF
 
 | | Structure of Get3d, a homolog of Get3, from Arabidopsis thaliana | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Manu, M.S, Barlow, A.N, Clemons Jr, W.M, Ramasamy, S. | | Deposit date: | 2022-09-23 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
8EGK
 
 | | Re-refinement of Crystal Structure of NosGet3d, the All4481 protein from Nostoc sp. PCC 7120 | | Descriptor: | AZIDE ION, All4481 protein | | Authors: | Barlow, A.N, Manu, M.S, Ramasamy, S, Clemons Jr, W.M. | | Deposit date: | 2022-09-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
