4HPP
 
 | | Crystal structure of novel glutamine synthase homolog | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Ladner, J.E, Atanasova, V, Dolezelova, Z, Parsons, J.F. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of PA5508, a Hexameric Glutamine Synthetase Homologue.
Biochemistry, 51, 2012
|
|
3LX2
 
 | |
3LX1
 
 | |
1NMO
 
 | | Structural genomics, protein ybgI, unknown function | | Descriptor: | FE (III) ION, Hypothetical protein ybgI | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1NMP
 
 | | Structural genomics, ybgI protein, unknown function | | Descriptor: | Hypothetical protein ybgI, MAGNESIUM ION | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
3R2Q
 
 | | Crystal Structure Analysis of yibF from E. Coli | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, PHOSPHATE ION, ... | | Authors: | Ladner, J.E, Harp, J, Schaab, M, Stournan, N.V, Armstrong, R.N. | | Deposit date: | 2011-03-14 | | Release date: | 2012-03-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural and Functional Genomics of YibF, a Glutathione Transferase Homologue from Escherichia coli
to be published
|
|
1R4W
 
 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
1RUV
 
 | | RIBONUCLEASE A-URIDINE VANADATE COMPLEX: HIGH RESOLUTION RESOLUTION X-RAY STRUCTURE (1.3 A) | | Descriptor: | RIBONUCLEASE A, TERTIARY-BUTYL ALCOHOL, URIDINE-2',3'-VANADATE | | Authors: | Ladner, J.E, Wladkowski, B, Svensson, L.A, Sjolin, L, Gilliland, G.L. | | Deposit date: | 1995-07-27 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray structure of a ribonuclease A-uridine vanadate complex at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1BHN
 
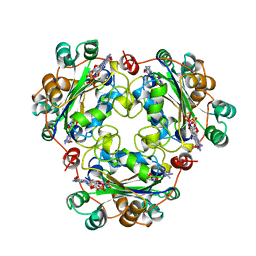 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM A FROM BOVINE RETINA | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-06-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of two isoforms of nucleoside diphosphate kinase from bovine retina.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2RGJ
 
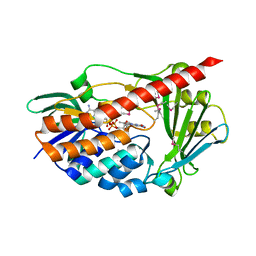 | | Crystal structure of flavin-containing monooxygenase PhzS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | Authors: | Ladner, J.E, Parsons, J.F, Greenhagen, B.T, Robinson, H. | | Deposit date: | 2007-10-03 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pyocyanin Biosynthetic Protein PhzS.
Biochemistry, 47, 2008
|
|
1MTC
 
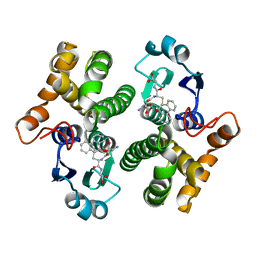 | | GLUTATHIONE TRANSFERASE MUTANT Y115F | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, Glutathione S-transferase YB1 | | Authors: | Ladner, J.E, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Local protein dynamics and catalysis: detection of segmental motion associated with rate-limiting product release by a glutathione transferase
Biochemistry, 41, 2002
|
|
3GX0
 
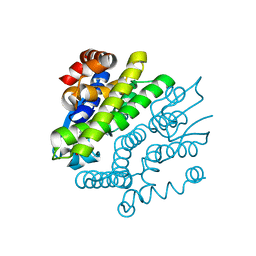 | | Crystal Structure of GSH-dependent Disulfide bond Oxidoreductase | | Descriptor: | GST-like protein yfcG, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Ladner, J.E, Harp, J.M, Wadington, M.C, Armstrong, R.N. | | Deposit date: | 2009-04-01 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase.
Biochemistry, 48, 2009
|
|
2QBV
 
 | | Crystal Structure of Intracellular Chorismate Mutase from Mycobacterium Tuberculosis | | Descriptor: | CHORISMATE MUTASE | | Authors: | Ladner, J.E, Reddy, P.T, Kim, S.K, Reddy, S.-K, Nelson, B.C. | | Deposit date: | 2007-06-18 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comparative biochemical and structural analysis of the intracellular chorismate mutase (Rv0948c) from Mycobacterium tuberculosis H(37)R(v) and the secreted chorismate mutase (y2828) from Yersinia pestis.
Febs J., 275, 2008
|
|
2GBB
 
 | | Crystal structure of secreted chorismate mutase from Yersinia pestis | | Descriptor: | CITRIC ACID, SULFATE ION, putative chorismate mutase | | Authors: | Ladner, J.E, Reddy, P.T, Nelson, B.C, Robinson, H, Kim, S.-K. | | Deposit date: | 2006-03-10 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A comparative biochemical and structural analysis of the intracellular chorismate mutase (Rv0948c) from Mycobacterium tuberculosis H(37)R(v) and the secreted chorismate mutase (y2828) from Yersinia pestis.
Febs J., 275, 2008
|
|
5DAI
 
 | |
5DA7
 
 | | monomeric PCNA bound to a small protein inhibitor | | Descriptor: | DNA polymerase sliding clamp 1, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Ladner, J.E, Altieri, A.S, Kelman, Z. | | Deposit date: | 2015-08-19 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A small protein inhibits proliferating cell nuclear antigen by breaking the DNA clamp.
Nucleic Acids Res., 44, 2016
|
|
2IJH
 
 | | Crystal structure analysis of ColE1 ROM mutant F14W | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IJJ
 
 | | Crystal structure analysis of ColE1 ROM mutant F14Y | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IP2
 
 | |
1BE4
 
 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM B FROM BOVINE RETINA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-05-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleoside diphosphate kinase from bovine retina: purification, subcellular localization, molecular cloning, and three-dimensional structure.
Biochemistry, 37, 1998
|
|
1DBF
 
 | |
1T9M
 
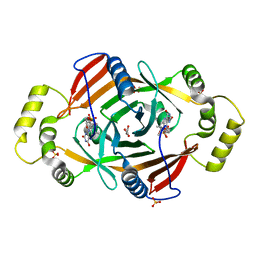 | | X-ray crystal structure of phzG from pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-05-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1K0E
 
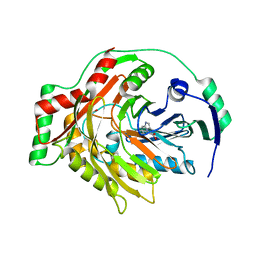 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM FORMATE GROWN CRYSTALS | | Descriptor: | FORMIC ACID, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
1K0G
 
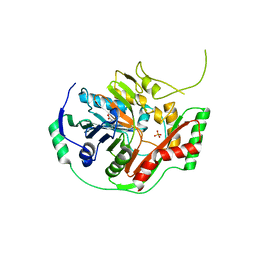 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM PHOSPHATE GROWN CRYSTALS | | Descriptor: | PHOSPHATE ION, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
4GRH
 
 | | Crystal structure of pabB of Stenotrophomonas maltophilia | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Bera, A, Atanasova, V, Ladner, J.E, Parsons, J.F. | | Deposit date: | 2012-08-24 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Aminodeoxychorismate Synthase from Stenotrophomonas maltophilia.
Biochemistry, 51, 2012
|
|
