4A7J
 
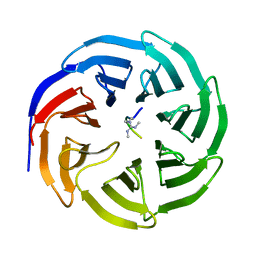 | | Symmetric Dimethylation of H3 Arginine 2 is a Novel Histone Mark that Supports Euchromatin Maintenance | | Descriptor: | HISTONE H3.1T, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Migliori, V, Muller, J, Phalke, S, Low, D, Bezzi, M, ChuenMok, W, Gunaratne, J, Capasso, P, Bassi, C, Cecatiello, V, DeMarco, A, Blackstock, W, Kuznetsov, V, Amati, B, Mapelli, M, Guccione, E. | | Deposit date: | 2011-11-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Dimethylation of H3R2 is a Newly Identified Histone Mark that Supports Euchromatin Maintenance
Nat.Struct.Mol.Biol., 19, 2012
|
|
6PQ1
 
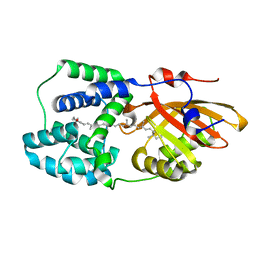 | | Structure of the Fremyella diplosiphon OCP1 | | Descriptor: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Dominguez-Martin, M.A, Bao, H, Kerfeld, C.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Comparative ultrafast spectroscopy and structural analysis of OCP1 and OCP2 from Tolypothrix.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
6PSH
 
 | |
6PSK
 
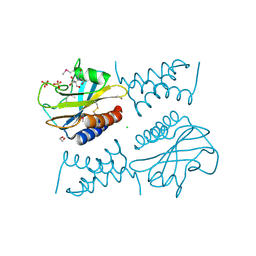 | | Crystal structure of the complex between periplasmic domains of antiholin RI and holin T from T4 phage, in P6522 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Antiholin, ... | | Authors: | Kuznetsov, V.B, Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-06-24 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of T4 Phage Lysis Control: DNA as the Signal for Lysis Inhibition.
J.Mol.Biol., 432, 2020
|
|
6PXE
 
 | |
6PX4
 
 | |
4H43
 
 | |
7AZT
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RE11660p | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S, Pandey, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AYV
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at cryogenic temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p, ... | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S. | | Deposit date: | 2020-11-13 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6T3L
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome in dark state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
6T3U
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome 1ps after photoexcitation | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
