2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2MLQ
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2D7H
 
 | | Crystal structure of the ccc complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*CP*CP*C)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7G
 
 | | Crystal structure of the aa complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7E
 
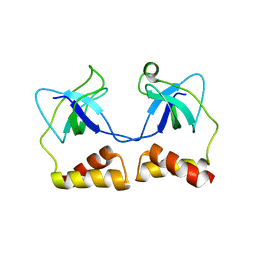 | | Crystal structure of N-terminal domain of PriA from E.coli | | Descriptor: | Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-18 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2UWM
 
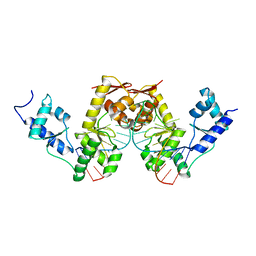 | | C-TERMINAL DOMAIN(WH2-WH4) OF ELONGATION FACTOR SELB IN COMPLEX WITH SECIS RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*GP*CP*CP*GP *GP*UP*CP*UP*GP*GP*CP*AP*AP*CP*GP*CP*C)-3', SELENOCYSTEINE-SPECIFIC ELONGATION FACTOR | | Authors: | Ose, T, Soler, N, Rasubala, L, Kuroki, K, Kohda, D, Fourmy, D, Yoshizawa, S, Maenaka, K. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Dynamic Interdomain Movement and RNA Recognition of the Selenocysteine-Specific Elongation Factor Selb.
Structure, 15, 2007
|
|
1OM2
 
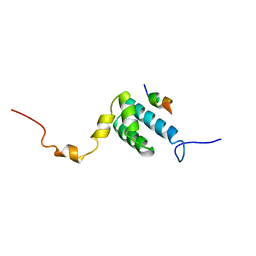 | | SOLUTION NMR STRUCTURE OF THE MITOCHONDRIAL PROTEIN IMPORT RECEPTOR TOM20 FROM RAT IN A COMPLEX WITH A PRESEQUENCE PEPTIDE DERIVED FROM RAT ALDEHYDE DEHYDROGENASE (ALDH) | | Descriptor: | PROTEIN (MITOCHONDRIAL ALDEHYDE DEHYDROGENASE), PROTEIN (MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20) | | Authors: | Abe, Y, Shodai, T, Muto, T, Mihara, K, Torii, H, Nishikawa, S, Endo, T, Kohda, D. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20.
Cell(Cambridge,Mass.), 100, 2000
|
|
1WSU
 
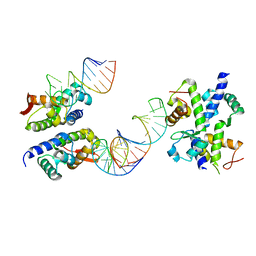 | | C-terminal domain of elongation factor selB complexed with SECIS RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*GP*CP*CP*GP*GP*UP*CP*U*GP*GP*CP*AP*AP*CP*GP*CP*C)-3', Selenocysteine-specific elongation factor | | Authors: | Yoshizawa, S, Rasubala, L, Ose, T, Kohda, D, Fourmy, D, Maenaka, K. | | Deposit date: | 2004-11-11 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for mRNA recognition by elongation factor SelB
Nat.Struct.Mol.Biol., 12, 2005
|
|
1HRE
 
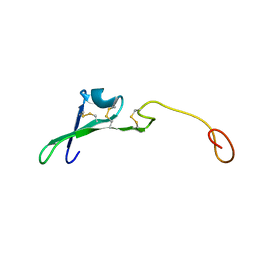 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1HRF
 
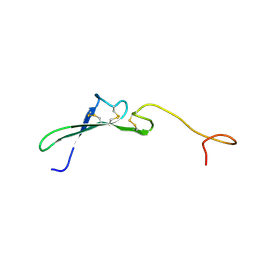 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
5YCQ
 
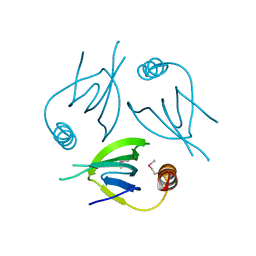 | | Unique Specificity-Enhancing Factor for the AAA+ Lon Protease | | Descriptor: | Heat shock protein HspQ | | Authors: | Abe, Y, Shioi, S, Kita, S, Nakata, H, Maenaka, K, Kohda, D, Katayama, T, Ueda, T. | | Deposit date: | 2017-09-08 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | X-ray crystal structure of Escherichia coli HspQ, a protein involved in the retardation of replication initiation
FEBS Lett., 591, 2017
|
|
1K4U
 
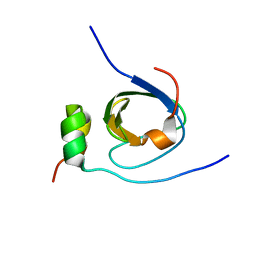 | | Solution structure of the C-terminal SH3 domain of p67phox complexed with the C-terminal tail region of p47phox | | Descriptor: | PHAGOCYTE NADPH OXIDASE SUBUNIT P47PHOX, PHAGOCYTE NADPH OXIDASE SUBUNIT P67PHOX | | Authors: | Kami, K, Takeya, R, Sumimoto, H, Kohda, D. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p.
EMBO J., 21, 2002
|
|
1L4V
 
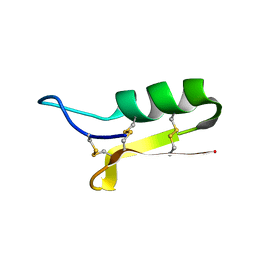 | | SOLUTION STRUCTURE OF SAPECIN | | Descriptor: | Sapecin | | Authors: | Hanzawa, H, Iwai, H, Takeuchi, K, Kuzuhara, T, Komano, H, Kohda, D, Inagaki, F, Natori, S, Arata, Y, Shimada, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H nuclear magnetic resonance study of the solution conformation of an antibacterial protein, sapecin.
FEBS Lett., 269, 1990
|
|
3AWR
 
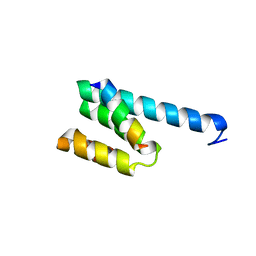 | |
3AX3
 
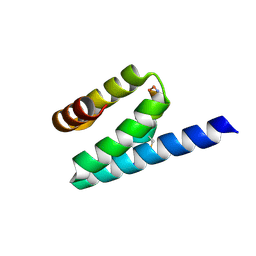 | |
3AX2
 
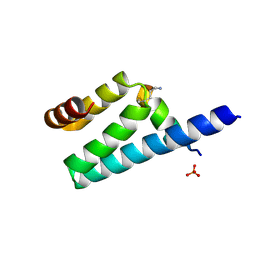 | | Crystal structure of rat TOM20-ALDH presequence complex: a disulfide-tethered complex with a non-optimized, long linker | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-28 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
3AX5
 
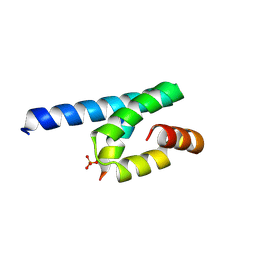 | | Crystal structure of rat TOM20-ALDH presequence complex: A complex (form1) between Tom20 and a disulfide-bridged presequence peptide containing D-Cys and L-Cys at the i and i+3 positions. | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
2D31
 
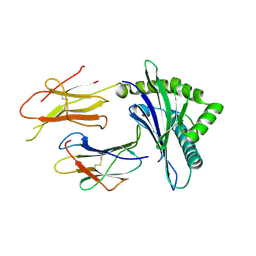 | | Crystal structure of disulfide-linked HLA-G dimer | | Descriptor: | 9-mer peptide from Histone H2A, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shiroishi, M, Kuroki, K, Ose, T, Rasubala, L, Shiratori, I, Arase, H, Tsumoto, K, Kumagai, I, Kohda, D, Maenaka, K. | | Deposit date: | 2005-09-23 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Efficient Leukocyte Ig-like Receptor Signaling and Crystal Structure of Disulfide-linked HLA-G Dimer
J.Biol.Chem., 281, 2006
|
|
2D3V
 
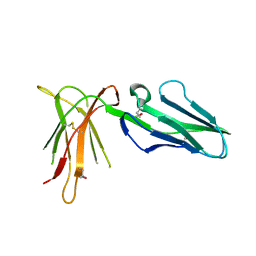 | | Crystal Structure of Leukocyte Ig-like Receptor A5 (LILRA5/LIR9/ILT11) | | Descriptor: | leukocyte immunoglobulin-like receptor subfamily A member 5 isoform 1 | | Authors: | Shiroishi, M, Kajikawa, M, Kuroki, K, Ose, T, Kohda, D, Maenaka, K. | | Deposit date: | 2005-10-03 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the human monocyte-activating receptor,
J.Biol.Chem., 281, 2006
|
|
2DWN
 
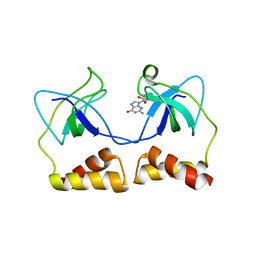 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWL
 
 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*(DC))-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWM
 
 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DYP
 
 | | Crystal Structure of LILRB2(LIR2/ILT4/CD85d) complexed with HLA-G | | Descriptor: | 9 Mer Peptide From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shiroishi, M, Kuroki, K, Rasubala, L, Kohda, D, Maenaka, K. | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d)
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2V1T
 
 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
2V1S
 
 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
