3P5H
 
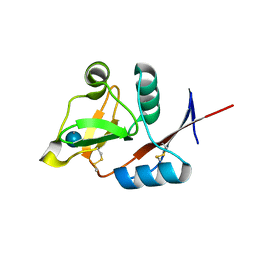 | | Structure of the carbohydrate-recognition domain of human Langerin with Laminaritriose | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, beta-D-glucopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6051 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5F
 
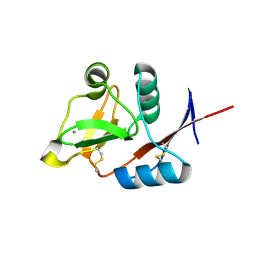 | | Structure of the carbohydrate-recognition domain of human Langerin with man2 (Man alpha1-2 Man) | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-D-mannopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7501 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5E
 
 | | Structure of the carbohydrate-recognition domain of human Langerin with man4 (Man alpha1-3(Man alpha1-6)Man alpha1-6Man) | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-D-mannopyranose | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7012 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5G
 
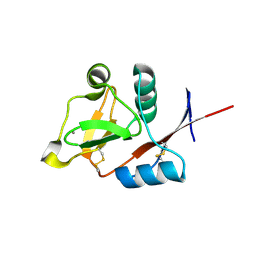 | | Structure of the carbohydrate-recognition domain of human Langerin with Blood group B trisaccharide (Gal alpha1-3(Fuc alpha1-2)Gal) | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-L-fucopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6027 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5I
 
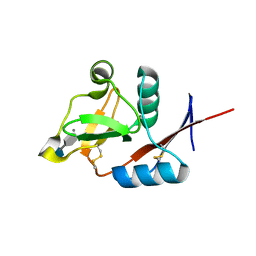 | | Structure of the carbohydrate-recognition domain of human Langerin with 6-SO4-Gal-GlcNAc | | Descriptor: | 6-O-sulfo-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 4 member K, CALCIUM ION | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3P5D
 
 | | Structure of the carbohydrate-recognition domain of human Langerin with man5 (Man alpha1-3(Man alpha1-6)Man alpha1-6)(Man- alpha1-3)Man | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-D-mannopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8013 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
4QNL
 
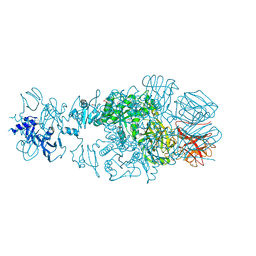 | | Crystal structure of tail fiber protein gp63.1 from E. coli phage G7C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Riccio, C, Browning, C, Prokhorov, N, Letarov, A, Leiman, P.G. | | Deposit date: | 2014-06-18 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Function of bacteriophage G7C esterase tailspike in host cell adsorption.
Mol. Microbiol., 105, 2017
|
|
4RU5
 
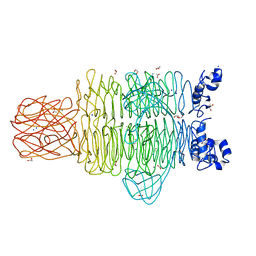 | | Crystal Structure of the Pseudomonas phage phi297 tailspike gp61 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Browning, C, Sycheva, L.V, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
4RU4
 
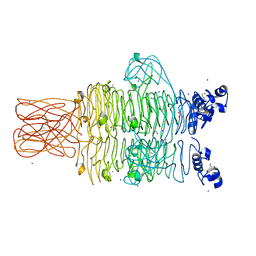 | | Crystal structure of the tailspike protein gp49 from Pseudomonas phage LKA1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Browning, C, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
8OQ0
 
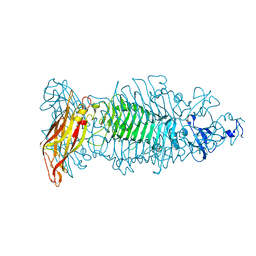 | | Crystal structure of tailspike depolymerase (APK09_gp48) from Acinetobacter phage APK09 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tailspike protein | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Popova, A.V, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii .
Int J Mol Sci, 24, 2023
|
|
8OPZ
 
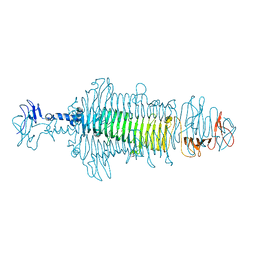 | | Crystal structure of a tailspike depolymerase (APK16_gp47) from Acinetobacter phage APK16 | | Descriptor: | GLYCEROL, Tailspike depolymerase (APK16_gp47) from Acinetobacter phage APK16 | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii .
Int J Mol Sci, 24, 2023
|
|
8OQ1
 
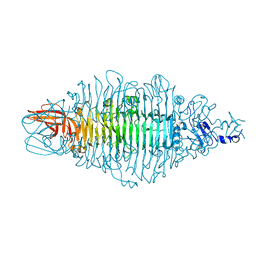 | | Crystal structure of tailspike depolymerase (APK14_gp49) from Acinetobacter phage vB_AbaP_APK14 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii.
Int J Mol Sci, 24, 2023
|
|
5W6S
 
 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) complex with Escherichia Coli O157-antigen | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Plattner, M, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2017-06-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
5W6F
 
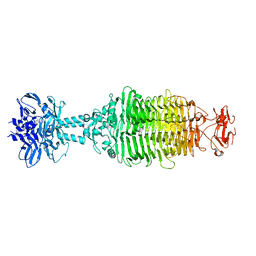 | |
5W6H
 
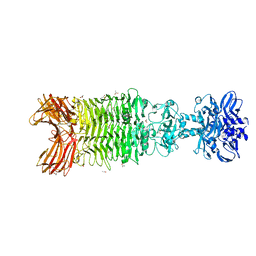 | | Crystal structure of Bacteriophage CBA120 tailspike protein 4 enzymatically active domain (TSP4dN, orf213) | | Descriptor: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Plattner, M, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2017-06-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
5W6P
 
 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, ZINC ION, ... | | Authors: | Plattner, M, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2017-06-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
