6EM6
 
 | |
6F14
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with isoquinoline and PKI (5-24) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOQUINOLINE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2017-11-21 | | Release date: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
6FJT
 
 | | 4-chloro-benzamidine in complex with thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-chloranylbenzenecarboximidamide, DIMETHYL SULFOXIDE, ... | | Authors: | Abazi, N, Heine, A, Klebe, G. | | Deposit date: | 2018-01-23 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | 4-chloro-benzamidine in complex with thrombin
To be published
|
|
6FFB
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant S205 - mutant Q148A - in complex with a nonsteroidal inhibitor | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bertoletti, N, Heine, A, Klebe, G, Marchais-Oberwinkler, S. | | Deposit date: | 2018-01-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational and structural studies uncover crucial amino acids determining activity and stability of 17 beta-HSD14.
J.Steroid Biochem.Mol.Biol., 189, 2019
|
|
6FPU
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex with 6-amino-2-((((3aS,5aR,8bS)-2,2,7,7-tetramethyltetrahydro-3aH-bis([1,3]dioxolo)[4,5-b:4',5'-d]pyran-3a-yl)methyl)amino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-azanyl-2-[[(1~{R},2~{S},6~{S},9~{R})-4,4,11,11-tetramethyl-3,5,7,10,12-pentaoxatricyclo[7.3.0.0^{2,6}]dodecan-6-yl]methylamino]-3,7-dihydroimidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2018-02-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Sugar Acetonides are a Superior Motif for Addressing the Large, Solvent-Exposed Ribose-33 Pocket of tRNA-Guanine Transglycosylase.
Chemistry, 24, 2018
|
|
6FSO
 
 | | Crystal Structure of TGT in complex with methyl({[5-(pyridin-3-yloxy)furan-2-yl]methyl})amine | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Fragments as Novel Starting Points for tRNA-Guanine Transglycosylase Inhibitors Found by Alternative Screening Strategies.
Chemmedchem, 15, 2020
|
|
6GBW
 
 | | Thrombin in complex with MI2100 ((S)-N-(2-(aminomethyl)-5-chlorobenzyl)-1-((benzylsulfonyl)-L-arginyl)pyrrolidine-2-carboxamide) | | Descriptor: | (2~{S})-~{N}-[[2-(aminomethyl)-5-chloranyl-phenyl]methyl]-1-[(2~{S})-5-carbamimidamido-2-[(phenylmethyl)sulfonylamino]pentanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2018-04-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
3MS3
 
 | | Crystal structure of Thermolysin in complex with Aniline | | Descriptor: | ANILINE, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3MSA
 
 | | Crystal structure of Thermolysin in complex with 3-Bromophenol | | Descriptor: | 3-bromophenol, CALCIUM ION, GLYCEROL, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3MSN
 
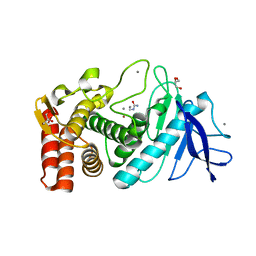 | | Crystal structure of Thermolysin in complex with N-methylurea | | Descriptor: | CALCIUM ION, GLYCEROL, N-METHYLUREA, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3MSF
 
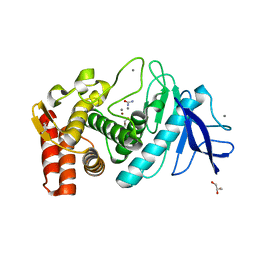 | | Crystal structure of Thermolysin in complex with Urea | | Descriptor: | CALCIUM ION, GLYCEROL, Thermolysin, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3N4A
 
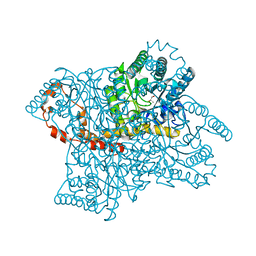 | | Crystal structure of D-Xylose Isomerase in complex with S-1,2-Propandiol | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, S-1,2-PROPANEDIOL, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3NN7
 
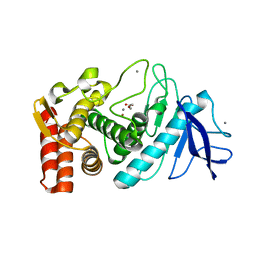 | | Crystal structure of Thermolysin in complex with 2-bromoacetate | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-06-23 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3N21
 
 | | Crystal structure of Thermolysin in complex with S-1,2-Propandiol | | Descriptor: | CALCIUM ION, S-1,2-PROPANEDIOL, Thermolysin, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-05-17 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3N9W
 
 | | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (IspD) in complex with 1,2-Propanediol | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, R-1,2-PROPANEDIOL, S-1,2-PROPANEDIOL | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-05-31 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3NX8
 
 | | human cAMP dependent protein kinase in complex with phenol | | Descriptor: | PHENOL, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-07-13 | | Release date: | 2011-07-13 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
7O04
 
 | |
7O6S
 
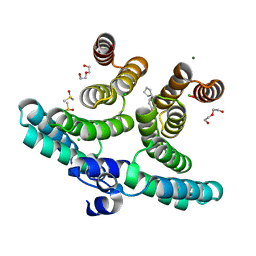 | | Crystal structure of a shortened IpgC variant in complex with N-(2H-1,3-benzodioxol-5-ylmethyl)cyclopentanamine | | Descriptor: | CHLORIDE ION, Chaperone protein IpgC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gardonyi, M, Heine, A, Klebe, G. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a shortened IpgC variant in complex with N-(2H-1,3-benzodioxol-5-ylmethyl)cyclopentanamine
To be published
|
|
7P42
 
 | |
7PE0
 
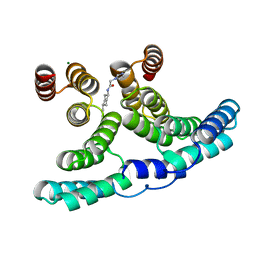 | | Crystal structure of IpgC in complex with J52 | | Descriptor: | 2-(1H-imidazol-1-yl)-N-(trans-4-methylcyclohexyl)acetamide, CHLORIDE ION, Chaperone protein IpgC, ... | | Authors: | Gardonyi, M, Heine, A, Klebe, G. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of IpgC in complex with J52
To be published
|
|
7PEF
 
 | |
7OWV
 
 | |
4Y35
 
 | |
4Y37
 
 | | Endothiapepsin in complex with fragement 305 | | Descriptor: | 4-chloro-2-methylthieno[2,3-d][1,2,3]diazaborinin-1(2H)-ol, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3A
 
 | | Endothiapepsin in complex with fragment 181 | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(3R,5S)-3,5-dimethylpiperidin-1-yl]methyl}-N,N-dimethyl-1,3,5-triazine-2,4-diamine, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
To Be Published
|
|
